Troubleshooting Synthetic Gene Circuits: Strategies for Stable Expression and Enhanced Reliability in Biomedical Applications
This article provides a comprehensive guide for researchers and drug development professionals tackling the persistent challenges of synthetic gene circuit expression and stability.
Troubleshooting Synthetic Gene Circuits: Strategies for Stable Expression and Enhanced Reliability in Biomedical Applications
Abstract
This article provides a comprehensive guide for researchers and drug development professionals tackling the persistent challenges of synthetic gene circuit expression and stability. Covering foundational principles to advanced applications, it explores the root causes of circuit failure, including metabolic burden, evolutionary instability, and host-circuit interactions. The content details innovative troubleshooting methodologies such as feedback controllers, memory circuits, and phase-separation techniques, alongside validation frameworks for assessing circuit longevity and performance. By synthesizing recent advances and comparative analyses of design strategies, this guide aims to equip scientists with the knowledge to build more robust and reliable genetic systems for therapeutic and diagnostic applications.
Understanding the Core Challenges: Why Synthetic Gene Circuits Fail
Metabolic Burden: Core Concepts and Definitions
What is metabolic burden in the context of synthetic gene circuits? Metabolic burden is the fitness cost imposed on a host cell by the expression of synthetic gene circuits. This occurs because the circuit actively consumes significant cellular resources—such as nucleotides, amino acids, energy (ATP), and enzymatic machinery—that the host cell creates for its own physiological functions, including growth and survival [1]. This resource competition can lead to reduced host growth rates and compromised circuit function.
What are the primary cellular resources that synthetic gene circuits consume? Synthetic gene circuits primarily consume the host's gene expression resources. This includes key components like [2]:
- Ribosomes (R): Essential for translating mRNA into protein.
- RNA polymerases: Required for transcribing DNA into mRNA.
- Energy (e): Cellular energy in the form of ATP and other anabolites needed to power transcription and translation.
- Amino acids and nucleotides: The fundamental building blocks for synthesizing proteins and nucleic acids.
How does metabolic burden ultimately lead to circuit failure? Burden creates a selective pressure that favors non-functional circuit mutants. Cells expressing the functional circuit experience a growth disadvantage because resources are diverted from their own essential processes [2]. Over time, faster-growing mutant cells that have acquired mutations disrupting circuit function (e.g., in promoters or coding sequences) will outcompete the original engineered cells. This evolutionary process eventually eliminates functional circuit from the population [2].
Diagnostic Guide: Identifying Metabolic Burden in Your Experiments
What are the key experimental indicators of high metabolic burden? The table below summarizes the primary quantitative and qualitative indicators of metabolic burden.
| Indicator | Description & Measurement | Typical Thresholds / Observations |
|---|---|---|
| Reduced Growth Rate | Slower cell division and prolonged culture doubling time compared to unengineered controls [2]. | Measured by optical density (OD) over time. A significant reduction (e.g., >20%) is a strong indicator. |
| Decreased Final Biomass | Lower saturation density in batch culture conditions [2]. | Measured as maximum OD. A lower yield suggests resources were diverted from biomass production. |
| Loss of Circuit Function Over Time | Decline in the population-level output of the circuit during prolonged culture (e.g., serial passaging) [2]. | Quantified by fluorescence (for reporters) or functional assays. A 50% reduction in output (τ50) is a common metric for failure [2]. |
| Increased Population Heterogeneity | Growing variability in circuit output between individual cells in a population [3]. | Observed via flow cytometry or microscopy; a wider distribution of expression levels suggests unstable control. |
How can I distinguish between metabolic burden and toxicity from my expressed protein? While both can reduce growth, they have distinct characteristics:
- Metabolic Burden: The negative effect on growth is proportional to the total level of synthetic gene expression, even for non-toxic proteins like GFP. Reducing expression levels (e.g., with a weaker promoter) should alleviate the growth defect [2].
- Protein Toxicity: The negative effect is specific to the function or misfolding of the protein itself. Growth defects will persist even at low expression levels if the protein is inherently toxic.
What modeling approaches can predict burden before experimental testing? "Host-aware" computational frameworks use ordinary differential equations to model host-circuit interactions. These models simulate the consumption of shared cellular resources (ribosomes, energy) by the circuit, dynamically coupling it to the host's growth rate. This allows for in silico prediction of burden and the evolutionary trajectory of the circuit population [2].
Mitigation Strategies: A Troubleshooting FAQ
What are the most effective design principles to minimize metabolic burden? The table below outlines key strategies supported by recent research.
| Strategy | Mechanism | Key Considerations |
|---|---|---|
| Implement Orthogonal Systems | Using genetic parts (e.g., bacterial transcription factors, phage recombinases) that interact weakly with the host's native networks [4]. | Reduces unintended cross-talk and interference with essential host processes [4]. |
| Employ Negative Feedback Control | The circuit's output protein regulates its own production, dampening overexpression and reducing resource consumption [2]. | Can extend the functional half-life (τ50) of the circuit. Post-transcriptional controllers (e.g., using sRNAs) can outperform transcriptional ones [2]. |
| Use Tunable Expression Systems | Systems like the DIAL (Distance-Induced Actuation of Levels) platform allow post-hoc adjustment of gene expression to find an optimal level that balances function and burden [3]. | Enables finding a "sweet spot" for gene expression that maintains function without over-burdening the host [3]. |
| Couple Circuit to Host Fitness | Artificially linking circuit function to an essential gene or survival mechanism [2]. | Makes mutations that disrupt the circuit also disadvantageous for survival, but can constrain circuit design [2]. |
How can I dynamically control expression to avoid burden? The DIAL system allows you to fine-tune expression after circuit delivery. It uses the distance between a promoter and gene—lengthened by a "spacer" sequence—to set a baseline expression level. Adding recombinase enzymes excises parts of the spacer, bringing the promoter closer and dialing expression up to predefined "high," "med," or "low" set points [3]. This enables real-time optimization to minimize burden while maintaining sufficient output.
My circuit function is still degrading rapidly. What advanced controllers can I use? For enhanced evolutionary longevity, consider multi-input controllers. These "host-aware" designs can use feedback based on both the circuit's output and the host's growth rate. Simulations show that growth-based feedback can extend the circuit's functional half-life more than threefold by directly countering the selective advantage of low-producing mutants [2].
Experimental Protocols for Burden Quantification
Protocol: Serial Passaging to Measure Circuit Longevity (Half-Life, τ50)
Objective: Quantify the evolutionary longevity of a synthetic gene circuit by measuring the time it takes for its population-level output to fall by 50% [2].
- Culture Inoculation: Start a batch culture of your engineered cells in a selective medium.
- Sampling and Dilution: Every 24 hours (or at your chosen interval), measure the culture's optical density (OD) and the circuit's output (e.g., fluorescence via flow cytometry). Dilute the culture into fresh medium to maintain continuous growth.
- Data Collection: Repeat Step 2 for at least 50-100 generations.
- Data Analysis:
Protocol: Measuring Single-Cell Burden with Flow Cytometry
Objective: Assess cell-to-cell heterogeneity and correlate circuit expression with growth markers at the single-cell level.
- Sample Preparation: Take samples from a growing culture of your circuit-bearing cells.
- Staining: Use a fluorescent dye (e.g., a membrane-permeable dye that binds nucleic acids) as a proxy for cellular biomass and growth rate.
- Flow Cytometry: Analyze the cells using a flow cytometer with appropriate lasers and filters to measure both the circuit reporter (e.g., GFP) and the growth marker dye simultaneously.
- Data Analysis: Create a scatter plot of circuit fluorescence versus growth marker fluorescence. A strong negative correlation indicates that high-expression cells have lower growth rates, directly demonstrating burden at the single-cell level.
Essential Visualizations
The Scientist's Toolkit: Research Reagent Solutions
| Research Reagent | Function & Application |
|---|---|
| Orthogonal Transcription Factors (e.g., bacterial TFs) | To build circuit integrators that minimize cross-talk with the host's native gene regulatory networks [4]. |
| Site-Specific Recombinases (e.g., Cre, from bacteriophage) | To implement permanent genetic memory or, as in the DIAL system, to tune expression levels by editing DNA spacer sequences [4] [3]. |
| CRISPR/Cas Components | To construct programmable synthetic gene circuits that can act as sensors, integrators, or actuators for endogenous genes without heavy reliance on host TFs [4]. |
| "Host-Aware" Modeling Software | A computational framework using ODEs to simulate host-circuit interactions, predict burden, and model population evolution before costly wet-lab experiments [2]. |
| Inducible Promoter Systems (e.g., Dexamethasone, β-Estradiol) | To provide precise temporal control over circuit activation, allowing researchers to separate growth phases from high-burden production phases [4]. |
| Small RNAs (sRNAs) | For post-transcriptional feedback control; can more effectively silence circuit mRNA and reduce burden compared to some transcriptional controllers [2]. |
Troubleshooting Guide: Common Circuit Failures & Solutions
This guide addresses the most frequent issues researchers encounter with the evolutionary stability of synthetic gene circuits.
Problem 1: Rapid Loss of Circuit Function Within Generations
- Symptoms: Circuit output (e.g., fluorescence, metabolite production) declines significantly over a small number of bacterial generations, often less than 50.
- Underlying Cause: The synthetic circuit imposes a metabolic burden on the host cell, diverting resources like ribosomes and amino acids away from host growth. This reduces the host's growth rate, creating a strong selective pressure for mutants where the circuit has been inactivated by mutation. These faster-growing mutants then outcompete the functional cells in the population [2] [5].
- Solution:
- Reduce Genetic Instability: Avoid repeated genetic sequences (e.g., homologous transcriptional terminators, repeated operator sequences) in your circuit design, as these are hotspots for recombination and deletion events [5].
- Lower Expression Level: Decrease the constitutive expression level of circuit genes. High expression levels are strongly correlated with low evolutionary half-life [5].
- Implement Negative Feedback: Design circuits that use negative feedback control to dynamically regulate expression, which can reduce burden and prolong functional output [2].
Problem 2: Inconsistent Performance Across Cell Populations
- Symptoms: Heterogeneous gene expression within a clonal population, leading to unreliable circuit behavior.
- Underlying Cause: Natural variation in cellular resources and stochastic gene expression, often exacerbated by competition for limited transcriptional and translational machinery [6] [3].
- Solution:
- Use Orthogonal Parts: Employ genetic parts (e.g., bacterial transcription factors, CRISPR/Cas components) that interact minimally with the host's native systems to reduce cross-talk and improve predictability [4].
- Implement Uniform Expression Control: Utilize systems like the DIAL (Dialable Expression) platform, which allows post-hoc adjustment of a circuit's expression set point to achieve uniform protein levels across a cell population [3].
Problem 3: Circuit Function is Unstable Under Dynamic Growth Conditions
- Symptoms: Circuit behavior changes unpredictably as host cell growth rates vary, for example, between different growth phases or media.
- Underlying Cause: Growth-mediated dilution: as cells grow and divide, circuit components like transcription factors are diluted, which can destabilize circuit dynamics and lead to failure [7] [8].
- Solution:
- Leverage Transcriptional Condensates: Engineer circuits where transcription factors are fused to intrinsically disordered regions (IDRs). These form droplet-like compartments via liquid-liquid phase separation, concentrating key components and buffering against dilution [8].
- Design for Growth-Feedback Robustness: Select circuit topologies that are known to maintain function, such as certain negative feedback loops or incoherent feed-forward loops, even when coupled with host growth [7].
Frequently Asked Questions (FAQs)
Q1: What are the key metrics for quantifying the evolutionary stability of my gene circuit?
Researchers typically use several metrics to measure evolutionary longevity [2]:
- P₀: The initial total circuit output of the ancestral population.
- τ₍±₁₀₎: The time (or number of generations) until the total population-wide output falls outside the range of P₀ ± 10%.
- τ₍₅₀₎ (Evolutionary Half-life): The time until the total population-wide output falls below 50% of P₀. This measures the "persistence" of circuit function.
Q2: My circuit needs high expression to be effective, but this makes it evolutionarily unstable. Are there design trade-offs?
Yes, this is a fundamental challenge. Quantitative studies show a direct trade-off: higher expression levels come at the cost of lower evolutionary stability [2] [5]. The table below summarizes experimental data showing how design choices impact evolutionary half-life.
Table 1: Impact of Circuit Design on Evolutionary Half-Life (Experimental Data)
| Circuit Design Feature | Impact on Evolutionary Half-life (τ₍₅₀₎) | Key Finding |
|---|---|---|
| High Expression Level | Decreased | A 4-fold increase in expression can reduce evolutionary half-life more than 17-fold [5]. |
| Removal of Repeated Sequences (e.g., homologous terminators) | Increased | Eliminating sequence homology between terminators can more than double the circuit's half-life [5]. |
| Use of Inducible Promoters | Increased | Circuits with inducible promoters show greater stability than those with constitutive promoters [5]. |
| Negative Autoregulation | Increased (Short-term) | Prolongs the duration of stable output in the short term [2]. |
| Growth-Based Feedback | Increased (Long-term) | Extends the functional half-life (τ₍₅₀₎) of the circuit [2]. |
| Post-transcriptional Control (sRNA) | Outperforms Transcriptional Control | Generally provides stronger control with reduced controller burden, enhancing longevity [2]. |
Q3: Can a gene circuit that has lost its function ever regain it through evolution?
Under specific selective pressures, yes, lost circuit function can be regained, though not always through direct reversion of the original mutation. Research in yeast has shown that broken circuits can adapt to restoring a beneficial function (e.g., drug resistance) through extracircuit mutations in the host genome. These mutations can elevate basal expression levels of the broken circuit or otherwise compensate for the lost function, rather than repairing the circuit's original coding sequence [9].
Experimental Protocols for Stability Testing
Protocol 1: Serial Passaging for Measuring Evolutionary Half-life
This is a standard method for quantifying how long a circuit remains functional in a growing microbial population.
- Starting Culture: Inoculate a clonal population of engineered cells into fresh, selective liquid medium.
- Growth and Dilution: Allow the culture to grow for a set period (e.g., to stationary phase, or for a fixed number of generations). Each day, perform a serial transfer by diluting the culture into fresh medium. A typical dilution factor allows for approximately 6-10 new generations per day [5].
- Monitor Output: At regular intervals (e.g., every 24 hours or 20 generations), sample the population.
- Induce the circuit if it uses an inducible promoter.
- Measure the population-level output (e.g., fluorescence via flow cytometry or plate reader, production of a specific metabolite).
- Plate samples to obtain single colonies and check for function at the single-cell level.
- Data Analysis: Plot the normalized circuit output against time (or number of generations). The evolutionary half-life (τ₍₅₀₎) is the time it takes for the output to fall to 50% of its initial value [2] [5].
Protocol 2: Identifying Loss-of-Function Mutations
When a circuit fails, identifying the mutation is crucial for redesign.
- Isolate Non-Functional Clones: From the evolved population, pick single colonies that show no or low circuit output.
- Sequence the Circuit: Isolate the plasmid (or amplify the genomic region) containing the gene circuit from the non-functional clones. Use Sanger sequencing or whole-plasmid sequencing to identify mutations.
- Common Mutations to Check [5]:
- Deletions between repeated sequences (promoters, terminators).
- Point mutations in promoters, ribosome binding sites (RBS), or coding sequences.
- Insertion sequence (IS) element insertions, often in the scar sequences between BioBrick parts.
- Verify Causality: Clone the identified mutant circuit back into a fresh host cell to confirm that the mutation alone causes the loss of function.
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Key Reagents for Engineering Evolutionary Robustness
| Research Reagent / Tool | Function in Circuit Design | Application in Stability Research |
|---|---|---|
| Orthogonal Transcription Factors (e.g., bacterial TFs in plants) [4] | Provides regulation that minimizes cross-talk with the host's native networks. | Reduces unintended interactions, making circuit performance more predictable and less likely to disrupt host fitness. |
| Small RNAs (sRNAs) [2] | Enables post-transcriptional regulation of circuit genes. | Generally outperforms transcriptional regulation in feedback controllers, offering strong control with lower burden. |
| Intrinsically Disordered Regions (IDRs) [8] | Fused to transcription factors to drive liquid-liquid phase separation. | Forms transcriptional condensates that buffer against growth-mediated dilution, stabilizing circuit memory and function. |
| Cre Recombinase (in DIAL system) [3] | Edits DNA spacer length between a promoter and gene. | Allows post-hoc fine-tuning of a circuit's expression set point in delivered cells for uniform, stable performance. |
| Serine Integrases/Recombinases (e.g., PhiC31, Bxb1, Flp) [10] | Catalyzes irreversible DNA recombination to create memory circuits. | Used to build logic gates and record past cellular events, though the logic is typically irreversible. |
| Host-Aware Computational Models [2] [7] | Multi-scale frameworks simulating host-circuit interactions, mutation, and competition. | Predicts evolutionary longevity in silico and evaluates controller architectures before costly experimental implementation. |
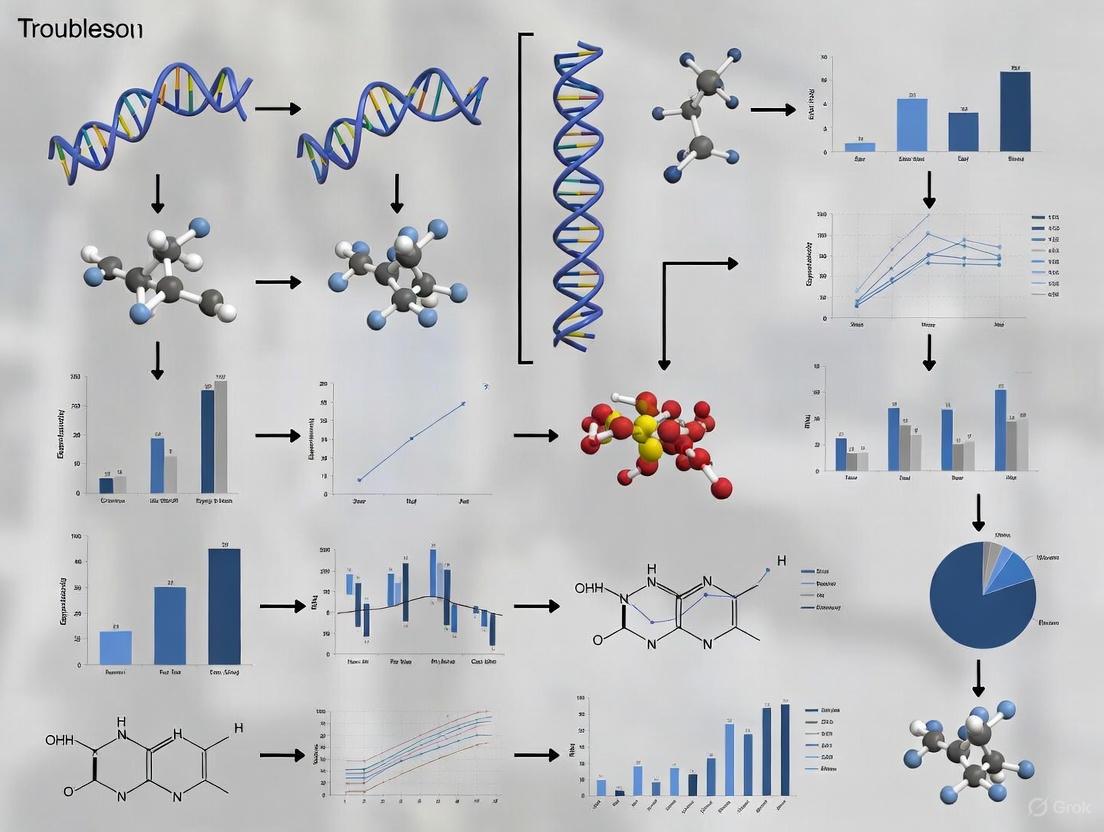
Visualizing Key Concepts and Workflows
Circuit Loss via Mutation & Selection
Burden-Feedback Loop
Core Concepts: Understanding Host-Circuit Interactions
What are host-circuit interactions? Synthetic gene circuits are not isolated entities; they function within a living cell and must utilize the host's native gene expression resources, such as ribosomes, amino acids, and RNA polymerases [2]. This sharing of resources creates an inherent interaction between the circuit and the host. The consumption of these cellular resources by the synthetic circuit disrupts the cell's natural homeostasis, a phenomenon often termed "metabolic burden" [4]. This burden frequently manifests as a reduction in cellular growth rate [2].
Why do these interactions cause problems? In microbes, growth rate is directly analogous to fitness. Therefore, a cell carrying a burdensome synthetic circuit is at a selective disadvantage compared to its unengineered or less-burdened counterparts [2]. During cell division, mutations can occur in the synthetic circuit. Mutations that reduce circuit function and, consequently, its resource consumption, provide a growth advantage to those cells. These "cheater" mutants can outcompete the original engineered cells, leading to the rapid evolutionary loss of circuit function in the population [2]. This is a primary reason why engineered circuits often lose functionality over time.
What is orthogonality and why is it important? Orthogonality is a fundamental design principle in synthetic biology. It refers to the use of genetic parts that interact strongly with each other but have minimal interaction with the host's natural cellular components [4]. This is often achieved by using parts derived from other organisms, such as bacterial transcription factors, phage-derived recombinases, or the CRISPR/Cas system [4]. Designing for orthogonality helps minimize unwanted cross-talk and reduces the metabolic burden on the host, thereby improving circuit stability and predictability [4].
Troubleshooting Common Problems
FAQ: My gene circuit's performance is declining rapidly over multiple cell generations. What is happening? This is a classic symptom of evolutionary instability. Your circuit is likely imposing a significant metabolic burden, creating a strong selection pressure for "cheater" mutants that have inactivating mutations in the circuit. These faster-growing mutants take over the population, causing a drop in the overall population-level output [2].
- Solution Strategies:
- Reduce Burden: Lower the expression level of your circuit components to the minimum required for function.
- Implement Genetic Controllers: Use feedback control systems that can sense and regulate circuit output or host growth to disincentivize the emergence of loss-of-function mutants [2].
- Couple to Essential Genes: Artificially link the function of your circuit to an essential gene required for host survival [2].
FAQ: The expression of my circuit is highly variable between cells, even in a clonal population. How can I fix this? Cell-to-cell variability (noise) can be caused by resource competition, stochastic binding of transcription factors, or mutations that alter the feedback dynamics of the circuit [11].
- Solution Strategies:
- Use Orthogonal Parts: Implement transcription factors and promoters from other species to avoid competition with host regulatory networks [4].
- Implement Feedback Loops: Negative feedback loops can suppress variation and stabilize expression levels [2].
- Stabilize with Transcriptional Condensates: Recent research shows that fusing transcription factors to intrinsically disordered regions (IDRs) can form condensates via liquid-liquid phase separation. These condensates concentrate circuit components, buffering against dilution and potentially stabilizing expression [8].
FAQ: My circuit works perfectly in one host strain but fails in another. Why? Different host strains can have varying genetic backgrounds, resource pools, and expression capacities. Your circuit may be interacting differently with these distinct cellular contexts.
- Solution Strategies:
- Characterize Parts in New Hosts: Always re-characterize your genetic parts (promoters, RBSs) in the specific host strain you plan to use.
- Host-Aware Modeling: Use computational models that account for host-circuit interactions, such as resource sharing and growth feedback, to predict circuit behavior in a new strain before experimental testing [2].
- Consider "Mid-Scale Evolution": If possible, subject your circuit to directed evolution in the new host strain under a relevant selection pressure to optimize its function in that specific context [11].
Experimental Protocols for Diagnosis & Mitigation
The table below outlines key experimental approaches for diagnosing and mitigating issues related to host-circuit interactions.
Table 1: Diagnostic and Mitigation Protocols for Host-Circuit Interactions
| Protocol Goal | Key Experimental Steps | Key Measurements & Outputs |
|---|---|---|
| Quantifying Evolutionary Longevity [2] | 1. Serial passaging of engineered population in batch culture.2. Regular sampling and measurement of population-level output (e.g., fluorescence).3. Genomic analysis of sampled populations to identify mutations. | - P₀: Initial output.- τ±10: Time until output deviates by >10% from P₀.- τ₅₀: Time until output falls below 50% of P₀. |
| Implementing Growth-Based Feedback [2] | 1. Design a controller that senses host growth rate or a proxy.2. Construct a circuit where this controller actuates repression of the synthetic gene (e.g., via sRNAs).3. Integrate the controller circuit and test in serial passaging experiments. | - Comparison of τ₅₀ and τ±10 between open-loop and closed-loop (controlled) circuits.- Measurement of reduced burden (improved growth rate) in functional state. |
| Stabilizing Circuits via Phase Separation [8] | 1. Fuse transcription factors (TFs) in your circuit to Intrinsically Disordered Regions (IDRs).2. Introduce fusion proteins into cells and confirm formation of transcriptional condensates via microscopy.3. Measure the stability of circuit output over time with and without IDR fusions. | - Visualization of condensates as bright, fluorescent foci.- Enhanced production yield in bioproduction pathways.- Increased resilience of circuit memory under dynamic growth. |
Research Reagent Solutions
Table 2: Essential Research Reagents and Their Functions
| Reagent / Tool Category | Specific Examples | Function in Troubleshooting Host-Circuit Interactions |
|---|---|---|
| Orthogonal Regulators | Bacterial TFs (e.g., TetR, LacI), CRISPR/Cas systems, Phage recombinases (e.g., Cre, Flp) [4] | Reduces cross-talk with host networks and decreases metabolic burden by using independent cellular machinery. |
| Inducible Promoters | Dexamethasone-, β-Estradiol-, Copper-, or Ethanol-responsive promoters [4] | Allows external control of circuit timing and expression level, enabling burden management and dynamic experiments. |
| Genetic Controllers | Negative autoregulation circuits, Growth-rate feedback controllers (using sRNAs for post-transcriptional control) [2] | Maintains circuit output and extends evolutionary longevity by automatically adjusting expression to mitigate burden. |
| Stabilization Tools | Transcription Factor-IDR fusions [8] | Forms condensates to protect circuit components from growth-mediated dilution, enhancing stability. |
| Modeling Frameworks | Host-aware ODE models, Multi-scale population models [2] | Predicts circuit behavior, burden, and evolutionary dynamics in silico before costly experimental implementation. |
Supporting Diagrams and Workflows
Diagram: Burden-Driven Evolutionary Failure of a Synthetic Gene Circuit
Diagram: Architecture of a Synthetic Gene Circuit with Controller
Growth-mediated dilution presents a fundamental challenge in synthetic biology, where the engineered gene circuits lose their functionality due to the dilution of key molecular components as host cells grow and divide. This phenomenon occurs because cell growth causes a global reduction in the concentrations of all circuit components, which can significantly destabilize circuit behavior and lead to complete functional collapse. Synthetic biology aims to program cells for useful tasks in medicine, biotechnology, and environmental engineering, but these genetic programs often fail because cell growth dilutes the key molecules needed to keep them running [8].
The problem is particularly acute in applications requiring long-term stability, such as industrial bioproduction where sustaining circuit activity during repeated culture dilutions is critical for minimizing inducer costs and ensuring consistent product yields [12]. Similarly, engineered probiotics for therapeutic applications must maintain reliable circuit performance under fluctuating nutrient conditions after ingestion [12]. Understanding and mitigating growth-mediated dilution is therefore essential for advancing synthetic biology applications from laboratory curiosities to real-world solutions.
Troubleshooting Guides
Problem Diagnosis Table
Table 1: Common Symptoms and Causes of Growth-Mediated Dilution Issues
| Observed Symptom | Potential Causes | Diagnostic Experiments |
|---|---|---|
| Circuit memory loss: Inability to maintain bistable 'ON' state after removal of initial stimulus [13] | Self-activation circuit topology; Rapid dilution of transcription factors during fast growth [12] [13] | Measure temporal dynamics of fluorescence and cell density after diluting activated cells into fresh medium [13] |
| Reduced bioproduction yield in prolonged cultures [12] | Dilution of key enzymes in metabolic pathways during scale-up [12] | Monitor product formation rates across different growth phases and dilution regimes |
| Inconsistent biosensor performance under dynamic growth conditions [12] | Growth-dependent variation in transcription factor concentrations [12] | Characterize dose-response curves at different growth rates |
| Loss of population-level synchrony in coordinated behaviors [13] | Growth-mediated dilution of signaling molecules or quorum sensing components [13] | Track single-cell expression distributions over multiple generations |
Circuit Failure Mechanisms and Signatures
Table 2: Quantitative Signatures of Growth-Mediated Circuit Failures
| Failure Mechanism | Dynamic Signature | Key Parameters Affected |
|---|---|---|
| Continuous response curve deformation [7] [14] | Gradual loss of adaptation precision and sensitivity; Altered input-output relationships [7] [14] | Reduced precision (final state deviation from basal); Decreased response sensitivity [14] |
| Induced or strengthened oscillations [7] [14] | Emergence of sustained oscillations not present in non-growth conditions [7] [14] | Oscillation amplitude and period modifications; Possible circuit reconfiguration to oscillatory regime [7] |
| Sudden switching to alternative attractors [7] [14] | Bistability loss; Memory circuit failure; Hysteresis collapse [7] [13] [14] | Loss of bistable range; Reduced hysteresis width; Inability to maintain predetermined states [13] |
Figure 1: Diagnostic Framework for Growth-Mediated Dilution Issues
Experimental Protocol: Validating Phase Separation Solution
Objective: To implement and validate a phase separation strategy for buffering growth-mediated dilution in a self-activation (SA) gene circuit.
Background: This protocol describes how to engineer transcriptional condensates by fusing intrinsically disordered regions (IDRs) to transcription factors, creating local concentrations that resist global dilution during cell growth [12] [15].
Materials:
- E. coli host strains (e.g., DH10B, ΔlacIΔaraCBAD mutant of MG1655)
- Plasmid constructs for SA circuit (e.g., OP174) and Drop-SA circuits (e.g., OP153 with FUSn, OP203 with RLP20)
- Inducers: L-(+)-Arabinose (Sigma A3256)
- FRAP-capable confocal microscope
- Flow cytometer
- Molecular biology reagents for cloning
Procedure:
Circuit Construction:
Transformation and Culture:
- Transform constructs into appropriate E. coli strains using standardized protocols (e.g., Zymo Research Mix & Go kit).
- Culture transformed cells in LB medium with appropriate antibiotics.
- Activate circuits by adding L-arabinose (0-10 mM range) during mid-exponential phase.
Dilution and Memory Testing:
- Dilute activated cells into fresh medium with varying L-arabinose concentrations.
- Monitor cell density (OD600) and fluorescence over time.
- Compare memory retention between SA and Drop-SA circuits.
Condensate Visualization and Validation:
- Image condensate formation using fluorescence microscopy.
- Perform FRAP (Fluorescence Recovery After Photobleaching) to confirm liquid-liquid phase separation properties [15].
- Analyze condensate dynamics and recovery kinetics.
Quantitative Analysis:
- Measure hysteresis properties through dose-response curves under different initial conditions.
- Quantify population heterogeneity using flow cytometry.
- Calculate local concentration factors within condensates.
Troubleshooting Notes:
- If condensates do not form, verify IDR fusion integrity and test different linker sequences.
- If memory improvement is suboptimal, tune expression levels and test alternative IDRs.
- Account for cell-to-cell variability by conducting sufficient replicates (≥1000 cells for microscopy) [15].
Research Reagent Solutions
Table 3: Essential Research Reagents for Addressing Growth-Mediated Dilution
| Reagent / Tool | Function / Application | Example Usage |
|---|---|---|
| Intrinsically Disordered Regions (IDRs) [12] [15] | Promote phase separation through multivalent interactions; Enable condensate formation | FUSn (FUS N-terminal domain) or RLP20 (resilin-like polypeptide) fused to transcription factors [15] |
| Self-Activation (SA) Circuit [13] | Model system for studying growth-mediated memory loss; Sensitive to dilution effects | Bicistronic circuit with AraC and GFP under PBAD promoter to test memory maintenance [13] |
| Toggle Switch Circuit [13] | Alternative topology refractory to growth-mediated dilution; Double-negative feedback motif | Comparative studies with SA circuits to identify topology-dependent resilience [13] |
| FRAP (Fluorescence Recovery After Photobleaching) [15] | Validate liquid-like properties of condensates; Measure dynamics and molecular mobility | Confirm phase separation in Drop-SA circuits; Typical recovery time ~10-11 minutes [15] |
| Growth Feedback Models [7] [14] | Computational frameworks to predict circuit-host interactions; Identify robust topologies | Systematic screening of 425+ circuit topologies for growth resilience; Parameter sampling (2×10⁵ trials) [7] [14] |
Frequently Asked Questions
Mechanism & Diagnosis
What exactly is growth-mediated dilution and why does it disrupt synthetic gene circuits?
Growth-mediated dilution refers to the reduction in intracellular concentration of synthetic gene circuit components that occurs as cells grow and divide. As cells increase in volume and undergo division, the molecular components of synthetic circuits (transcription factors, enzymes, signaling molecules) become distributed throughout the larger cellular volume and among daughter cells, effectively reducing their concentrations [12]. This global reduction in component concentrations can significantly destabilize circuit behavior, particularly for circuits that rely on precise concentration thresholds for proper function, such as bistable switches and oscillators [12] [13].
How can I determine if growth-mediated dilution is causing my circuit failures?
Several diagnostic approaches can help identify growth-mediated dilution as the root cause:
- Monitor the temporal dynamics of circuit components during growth phases: rapid decline in expression levels during exponential growth followed by recovery in stationary phase suggests dilution effects [13].
- Compare circuit performance in fast-growing versus slow-growing conditions: growth-mediated issues will show condition-dependent failures [7].
- Test circuit memory by activating circuits then diluting into fresh medium: inability to maintain state indicates dilution problems [13].
- Use control circuits with different topologies known to have varying sensitivity to growth feedback (e.g., compare self-activation vs. toggle switches) [13].
Are certain circuit topologies more vulnerable to growth-mediated dilution?
Yes, circuit topology significantly influences vulnerability to growth-mediated dilution. Self-activation circuits implementing positive autoregulation are particularly sensitive to dilution effects and quickly lose memory function during rapid growth [13]. In contrast, toggle switches with double-negative feedback motifs are more refractory to growth-mediated dilution and can maintain memory better under dynamic growth conditions [13]. Systematic studies of 425 adaptive circuit topologies revealed that only a small subset maintains optimal performance under growth feedback, highlighting the importance of topology selection [7] [14].
Solutions & Implementation
What is the phase separation strategy for combating growth-mediated dilution?
The phase separation strategy involves engineering biomolecular condensates that locally concentrate transcription factors at promoter regions, creating protected microenvironments that resist global dilution [12] [8]. This is achieved by fusing intrinsically disordered regions (IDRs) to transcription factors, enabling them to form liquid-like droplets through liquid-liquid phase separation [12] [15]. These condensates maintain high local concentrations of key circuit components even as average cellular concentrations decrease during growth, thereby preserving circuit function [12] [15].
Figure 2: Mechanism of Phase Separation in Countering Growth-Mediated Dilution
What specific IDRs have been successfully used to buffer against growth-mediated dilution?
Two main types of IDRs have been experimentally validated for this application:
- FUSn: The N-terminal intrinsically disordered domain of the Fused in Sarcoma protein, a natural IDR known to promote liquid-liquid phase separation through multivalent interactions [15].
- RLP20: A synthetic resilin-like polypeptide engineered to drive biomolecular condensate formation with tunable properties [15]. Both have been successfully fused to transcription factors (AraC) in self-activation circuits, forming condensates at polar regions in E. coli and maintaining circuit memory under growth conditions that disrupt standard circuits [15].
How do I validate that my engineered condensates are functioning properly?
Several validation methods can confirm proper condensate function:
- Fluorescence Microscopy: Visualize condensate formation as bright, concentrated foci, typically at polar regions in bacteria [15].
- FRAP Analysis: Photobleach condensates and monitor fluorescence recovery, with typical recovery times of ~10-11 minutes confirming liquid-like properties [15].
- Hysteresis Testing: Compare dose-response curves under different initial conditions to confirm memory preservation in Drop-SA circuits [15].
- Single-Cell Analysis: Use flow cytometry to quantify population heterogeneity and identify cells with successful condensate formation [15].
Are there alternative strategies beyond phase separation for mitigating growth-mediated dilution?
Yes, several complementary strategies exist:
- Circuit Topology Engineering: Selecting naturally resilient topologies (e.g., toggle switches over self-activation circuits) [13].
- Growth Feedback Controllers: Implementing genetic controllers that monitor and regulate circuit function based on growth signals [2].
- Post-transcriptional Regulation: Using small RNAs for control, which may provide amplification enabling strong regulation with reduced burden [2].
- Host-Aware Design: Using computational frameworks that account for circuit-host interactions during the design process [2].
- Negative Autoregulation: Implementing feedback loops that can prolong short-term circuit performance [2].
Applications & Optimization
In which applications is addressing growth-mediated dilution most critical?
Growth-mediated dilution mitigation is particularly important for:
- Industrial Bioproduction: Where sustaining circuit activity during repeated culture dilutions for scale-up is critical for minimizing inducer costs and ensuring consistent product yields [12].
- Engineered Probiotics and Microbiome Therapeutics: Where therapeutic microbes must grow and divide under fluctuating nutrient conditions after ingestion while maintaining circuit function [12].
- Long-term Biosensing: Where reliable circuit performance under dynamically changing growth environments is essential for consistent detection signals [12].
- Cancer Therapeutics: Where engineered bacterial therapies must maintain function through multiple growth cycles in dynamic tumor environments [12].
What are the key metrics for evaluating solutions to growth-mediated dilution?
Several quantitative metrics can evaluate mitigation strategies:
- Memory Retention: Ability to maintain bistable states through growth phases [15] [13].
- Hysteresis Range: Width of bistable region in dose-response curves [15].
- Functional Half-life: Time for circuit output to fall by 50% (τ50) [2].
- Stability Duration: Time circuit maintains performance within ±10% of initial (τ±10) [2].
- Production Yield: Total output of desired products in bioproduction applications [12] [8].
- Precision and Sensitivity: Maintenance of adaptation properties in responsive circuits [7] [14].
How scalable is the phase separation approach for complex genetic circuits?
Current evidence suggests phase separation is a promising and potentially generalizable strategy [12] [8]. The minimal modification required (adding IDR fusions to key transcription factors) makes it applicable to various circuit designs without complete redesign [12]. Research demonstrates successful application in both simple self-activation circuits and more complex systems like cinnamic acid biosynthesis pathways [12] [8]. However, optimal implementation may require balancing condensate properties with circuit function and careful selection of IDR-cargo combinations to avoid unintended interactions [15].
Advanced Designs and Systems for Robust Circuit Function
Troubleshooting Guide: FAQs and Solutions for the DIAL System
This guide provides targeted troubleshooting for researchers implementing the DIAL (Dialable) promoter system to achieve precise, heritable set-points of transgene expression.
System Setup and Initial Transfection
Q: After transfection, my flow cytometry shows a completely bimodal expression profile (ON and OFF populations) instead of a uniform unimodal peak. What went wrong?
- Potential Cause 1: The synthetic Zinc Finger Activator (ZFa) is too strong. The DIAL system is designed for uniform, unimodal expression, but certain powerful transactivation domains can overwhelm this, leading to bistability and bimodality [16].
- Solution: Titrate the amount of ZFa plasmid used in transfection or switch to a ZFa with a weaker transactivation domain (e.g., from the COMET toolkit) [16]. The system should be characterized with ZFas that generate unimodal setpoints for reliable control [16].
- Potential Cause 2: Inefficient or uneven transfection. A bimodal distribution can simply mean that only a fraction of the cells received the genetic circuit.
- Solution:
- Always use a co-transfection marker (e.g., a constitutively expressed fluorescent protein on a separate plasmid) to gate and analyze only the successfully transfected cell population during flow cytometry [16].
- Optimize your transfection protocol for your specific cell line to improve efficiency and consistency. Consider using lentiviral delivery for more stable integration and uniform uptake, especially in primary cells and iPSCs [16].
Q: I am observing high background expression even in the absence of the ZFa ("OFF" state is not off).
- Potential Cause: Promoter leakiness. The minimal core promoter or the spacer sequence itself may have low-level basal activity.
- Solution:
- Verify the specificity of your ZFa and its binding sites. Ensure the ZFa binding sites are fully orthogonal to the host cell's native transcription factors [16].
- Test different minimal promoters or adjust the spacer sequence to minimize background activity. The DIAL framework is modular and allows for such optimizations [16].
Recombinase-Mediated Editing
Q: I've added Cre recombinase, but the shift to a higher expression setpoint is inefficient or incomplete.
- Potential Cause 1: Low recombinase activity or delivery issue. The Cre enzyme may not be active enough or may not have reached all cells.
- Solution:
- Validate Cre recombinase activity using a separate reporter cell line (e.g., a tdTomato-to-GFP switch reporter).
- Ensure you are using a reliable delivery method for Cre (e.g., high-efficiency transfection, cell-penetrating Cre protein, or adenoviral delivery).
- Confirm successful spacer excision by performing genotyping PCR on the extracted circuit DNA from your cell population. A successful edit will show a shorter PCR band corresponding to the excised promoter [16].
- Potential Cause 2: The spacer length is too long. While longer spacers increase the dynamic range, they might also make the initial state too repressed, and the edited state might not reach the desired absolute level [16].
- Solution: Consult the table below and consider using a DIAL promoter with a shorter spacer length for a more robust fold-change that meets your experimental needs.
Q: How do I design a DIAL system with more than two setpoints (e.g., Low, Med, High, Off)?
- Solution: Implement a nested DIAL promoter architecture. This involves incorporating multiple, orthogonal recombinase sites (e.g., loxP, FRT for Flp recombinase) within the spacer. By using different recombinases sequentially or in combination, you can create a series of progressively shorter spacers, thereby generating multiple discrete setpoints from a single promoter [3] [16].
Expression Stability and Long-Term Performance
Q: My carefully set expression level drifts down over multiple cell divisions. How can I maintain a stable setpoint?
- Potential Cause: Evolutionary burden and dilution. Cells expressing a high level of a non-essential transgene experience a growth disadvantage. Over time, faster-growing mutants with reduced expression (e.g., due to promoter mutations) will outcompete the ancestral engineered cells [2].
- Solution 1: Leverage the heritability of DIAL setpoints. A key advantage of the DIAL system is that the promoter edit is genetically encoded and stable. Once a setpoint is established via recombinase action, it is maintained heritably in daughter cells without the need for continuous input [16]. Ensure you are using a delivery method (like lentivirus) that promotes genomic integration for long-term stability.
- Solution 2: Integrate stability-enhancing genetic controllers. For very long-term cultures, consider implementing feedback circuits that couple essential host functions to your transgene. "Host-aware" designs, such as negative autoregulation or growth-based feedback, can theoretically improve evolutionary longevity by reducing the selective advantage of loss-of-function mutants [2].
- Solution 3: Explore biomolecular condensates. Emerging strategies involve fusing transcription factors to intrinsically disordered regions (IDRs) to drive liquid-liquid phase separation. These transcriptional condensates form at the promoter and can buffer against growth-mediated dilution, helping to maintain consistent expression levels across dynamic growth conditions [8].
Q: The expression in my cell population is highly variable, making it difficult to map levels to a phenotypic output.
- Potential Cause: The setpoint itself is not unimodal. This returns to the initial issue of using an appropriate ZFa and ensuring uniform delivery. For mapping gene dosage to phenotypes, a unimodal, uniform population response is critical [16].
- Solution:
- Re-optimize the ZFa strength and transfection as described in the first FAQ.
- Use fluorescence-activated cell sorting (FACS) to isolate a cell population with a very narrow range of expression after the setpoint has been established. This will create a more homogeneous starting population for your phenotype assays.
The Scientist's Toolkit: Research Reagent Solutions
| Item | Function / Explanation |
|---|---|
| Synthetic Zinc Finger Activators (ZFas) | Engineered transcription factors that bind orthogonal DNA sequences upstream of the minimal promoter. Different ZFa strengths (e.g., by varying the transactivation domain) allow for tuning the system's maximum output [16]. |
| DIAL Promoter Construct | The core genetic component containing tessellated ZFa binding sites, an excisable "spacer" sequence, and a minimal core promoter (e.g., TATA). Spacer length is a key tunable parameter [16]. |
| Cre Recombinase (and other site-specific recombinases) | Enzyme that catalyzes the excision of the DNA spacer flanked by its recognition sites (e.g., loxP), bringing the promoter closer to the transcription start site and shifting expression to a higher setpoint [3] [16]. |
| Lentiviral Delivery System | A method for stably integrating the DIAL circuit into the genome of hard-to-transfect cells, such as primary cells and induced pluripotent stem cells (iPSCs) [16]. |
| Co-transfection Marker | A fluorescent protein expressed from a separate, constitutive promoter. It allows for identifying and gating on successfully transfected cells during flow cytometry analysis, ensuring you only analyze cells that received the circuit [16]. |
Quantitative Data for DIAL System Design
Table 1: Impact of Spacer Length on DIAL Promoter Output
| Spacer Length (base pairs) | Relative Expression (Pre-Excision) | Fold Change (Post- vs. Pre-Excision) |
|---|---|---|
| 27 bp | High | Low |
| 203 bp | Medium | Medium |
| 263 bp | Low | High |
This table summarizes the tunability of the DIAL system. Increasing the length of the excisable spacer sequence decreases the initial ("Low" setpoint) expression level and correspondingly increases the fold-change achieved after Cre-mediated excision [16].
Table 2: Comparison of Stability-Enhancing Strategies
| Strategy | Mechanism | Key Advantage | Key Disadvantage |
|---|---|---|---|
| DIAL (Heritable Editing) | Genetically encoded, permanent promoter shortening after a one-time recombinase input [16]. | Stable, set-and-forget control; no ongoing resource drain on the cell. | Setpoints are fixed and not easily reversible. |
| Transcriptional Feedback | Negative autoregulation senses and adjusts circuit output [2]. | Can improve short-term stability and reduce burden. | Controller itself consumes resources; can be evolutionarily disrupted. |
| Phase Separation (Condensates) | Concentrates transcriptional machinery via liquid-liquid phase separation to buffer against dilution [8]. | A physical principle that enhances robustness under dynamic growth. | A nascent technology; requires fusion of specific protein domains (IDRs). |
Experimental Workflow and Mechanism
The following diagram illustrates the core mechanism of the DIAL system and a general workflow for its implementation and troubleshooting.
DIAL System Mechanism and Workflow
Frequently Asked Questions (FAQs) and Troubleshooting Guides
FAQ 1: What are the primary benefits of using negative autoregulation in a gene circuit?
Negative autoregulation (NAR), where a transcription factor represses its own promoter, provides two key advantages for synthetic gene circuits:
- Linearized Dose-Response: It can transform a steep, sigmoidal (non-linear) input-output dose-response into a linear one. This allows for more precise and graded control of gene expression over a wide range of inducer concentrations [17].
- Reduced Expression Noise: It significantly reduces cell-to-cell variability (noise) in gene expression. Experiments in yeast showed up to a 7-fold reduction in noise at intermediate induction levels, leading to a more uniform population response [17].
FAQ 2: My gene circuit's output is declining over multiple cell generations. What could be causing this?
This is a classic sign of evolutionary instability. Circuits that impose a high metabolic burden on the host cell slow its growth. Over time, faster-growing mutant cells that have lost or impaired circuit function will outcompete the original engineered cells [2]. This is a fundamental challenge in synthetic biology.
FAQ 3: What design strategies can I use to make my gene circuit more evolutionarily stable?
Controller architectures that use feedback to minimize burden can significantly extend functional longevity. Computational and experimental studies suggest:
- For Short-Term Performance: Negative autoregulation can help maintain function initially [2].
- For Long-Term Persistence: Growth-based feedback controllers, which sense and respond to the host's growth rate, can outperform other types. Post-transcriptional controllers using small RNAs (sRNAs) can also provide stronger control with less burden than transcriptional controllers [2].
- Multi-Input Controllers: Designs that combine different feedback mechanisms (e.g., sensing both circuit output and growth rate) can improve both short-term and long-term performance [2].
FAQ 4: My circuit is exhibiting high cell-to-cell variability (noise). How can I troubleshoot this?
High noise can stem from several sources. The table below outlines common causes and solutions.
| Cause | Troubleshooting Action |
|---|---|
| Low or Insufficient Sample | Ensure sample volume is at least 10 µL for fragment analysis. For sequencing samples cleaned with BigDye XTerminator, use a minimum of 65 µL [18]. |
| Old or Expired Reagents | Replace expired cartridges, cathode buffer containers, or other reagents before re-running samples [18]. |
| Sample Degradation | Limit the time samples are stored on-instrument. For long runs, limit plates to 48 samples. Ensure Hi-Di Formamide is less than a year old and has undergone fewer than 8 freeze-thaw cycles [18]. |
| Incorrect Dye Calibration | Perform a new spectral calibration using matrix standards and verify the correct dye set is selected in the plate setup software [18]. |
| Fundamental Circuit Design | Consider redesigning your circuit to include negative autoregulation, a known motif for noise reduction [17] [19]. |
FAQ 5: How does growth feedback generally affect gene circuit function?
Growth feedback is a major circuit-host interaction where the circuit affects cell growth, and growth in turn affects gene expression by diluting cellular components. A systematic study of over 400 adaptation circuits found it can cause failure through three main mechanisms [7]:
- Continuous Deformation: The response curve becomes distorted.
- Induced Oscillations: The circuit begins to oscillate unpredictably.
- Sudden Switching: The circuit jumps to a different, unintended stable state. Despite these generally negative effects, a small subset of circuit topologies is naturally robust to growth feedback [7].
Quantitative Data and Design Principles
Table 1: Performance Comparison of Genetic Controller Architectures [2]
| Controller Architecture | Key Feature | Short-Term Performance (τ±10) | Long-Term Persistence (τ50) | Key Advantage |
|---|---|---|---|---|
| Open-Loop (No Control) | No feedback | Low | Low | Baseline for comparison |
| Intra-Circuit Feedback | Negative autoregulation of circuit genes | High | Medium | Good initial performance |
| Growth-Based Feedback | Actuation based on host growth rate | Medium | High | Best long-term circuit survival |
| Post-Transcriptional Control | Uses sRNAs for silencing | Varies | High | Strong control with low burden |
Design Principle: Speeding Up Circuit Response Time The response time of a simple gene expression system is determined by the protein degradation rate (γ). A faster response can be achieved by destabilizing the protein (increasing γ). To maintain the same steady-state concentration, the production rate (β) must be increased proportionally. This creates a futile cycle but pays off in speed [19]. Negative autoregulation is a network motif that also accelerates the turn-on time of a gene's expression [19].
Experimental Protocols
Protocol 1: Implementing a Negative Autoregulatory "Linearizer" Circuit
This protocol is based on the construction and validation of a linearizer circuit in yeast [17].
- Circuit Design: Replace the constitutive promoter driving your transcription factor (e.g., TetR) with a promoter that the TF itself can repress (e.g., PGAL1-D12 for TetR). This creates a negative feedback loop on the regulator.
- Chromosomal Integration: Integrate the regulator and reporter (e.g., yEGFP) parts separately into the host chromosome to minimize copy number variability.
- Induction Curve Measurement:
- Culture cells harboring the circuit across a wide range of inducer concentrations (e.g., 0-100 ng/mL anhydrotetracycline (ATc)).
- Use flow cytometry to measure the population mean and distribution of the reporter fluorescence at each concentration.
- Data Analysis:
- Plot the mean fluorescence against the inducer concentration.
- A successful linearizer circuit will show a linear relationship (R² ≈ 0.99) from no induction up to ~90% saturation, unlike the sigmoidal curve of a non-autoregulatory cascade.
- The fluorescence distribution histograms should remain unimodal and narrow across all concentrations, indicating low noise.
Protocol 2: In Silico Modeling of Circuit Evolution with Growth Feedback
This protocol uses a multi-scale computational framework to predict circuit longevity [2].
- Model Formulation:
- Develop a set of ordinary differential equations (ODEs) that describe host-circuit interactions, including resource consumption (ribosomes, amino acids) and its impact on cellular growth rate.
- Key variables are often in molecules per cell.
- Define Mutation Scheme:
- Implement a state-transition model with several "mutation states" (e.g., 100%, 67%, 33%, 0% of nominal expression).
- Set transition rates so that function-reducing mutations are more likely.
- Simulate Population Dynamics:
- Run the model in repeated batch conditions (nutrient replenishment every 24 hours).
- Track the competition between different mutant strains and the total population-level output over time.
- Quantify Longevity:
- P₀: Record the initial total output.
- τ±10: Calculate the time for the total output to fall outside P₀ ± 10%.
- τ₅₀: Calculate the time for the total output to fall below P₀/2.
Key Signaling Pathways and Workflows
Diagram 1: Negative Autoregulation with Growth Feedback This diagram illustrates the core logic of a negatively autoregulated circuit operating within a host cell. The transcription factor (TF) represses both its own gene and the output gene. The resulting output protein can impose a metabolic burden, reducing the host's growth rate. The growth rate, in turn, feeds back into the system by diluting all cellular proteins, creating a complex interaction that impacts circuit stability and longevity [17] [7] [2].
Diagram 2: Bistable Toggle Switch States The genetic toggle switch is a classic bistable system with two stable states (A and B). The intermediate state is unstable; without external control, stochastic fluctuations quickly push cells into one of the two stable attractors. Real-time feedback control can be used to maintain the population in this unstable state [20].
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents and Tools for Genetic Controller Experiments
| Item | Function in Experiment | Example/Note |
|---|---|---|
| Anhydrotetracycline (ATc) | Small-molecule inducer; binds to and inactivates the TetR repressor, allowing gene expression. | Used in TetR-based systems (e.g., [17] [20]). |
| IPTG | Small-molecule inducer; binds to and inactivates the LacI repressor, allowing gene expression. | Used in LacI-based systems (e.g., [20]). |
| Cre Recombinase | Enzyme that catalyzes site-specific recombination of DNA between two loxP sites. | Used in the DIAL system to edit expression setpoints by excising DNA spacers [3]. |
| Microfluidic Device | Allows for long-term, single-cell imaging and dynamic control of the cellular environment. | Critical for real-time feedback control experiments (e.g., [20]). |
| Flow Cytometer | Measures fluorescence of individual cells in a population, enabling quantification of mean expression and noise. | Essential for characterizing dose-response and cell-to-cell variability [17]. |
| Spectral Calibration Standards | Used to calibrate fluorescent dye detection systems, ensuring accurate signal measurement. | Necessary for troubleshooting pull-up/pull-down artifacts in data [18]. |
| BigDye XTerminator Kit | Reagent for purifying sequencing reactions to remove unincorporated terminators. | Insufficient cleanup can cause low signal or dye blobs [18]. |
| Hi-Di Formamide | Used for sample denaturation before capillary electrophoresis sequencing. | Age and freeze-thaw cycles can degrade sample resolution [18]. |
This technical support center is designed to assist researchers and scientists in troubleshooting common issues encountered when working with recombinase and CRISPR-based synthetic gene circuits for memory and logic operations. The guidance is framed within the broader thesis of troubleshooting synthetic gene circuit expression and stability, addressing specific experimental challenges related to genetic instability, burden, and unpredictable performance to enhance the reliability of your research and drug development applications.
Frequently Asked Questions (FAQs)
FAQ 1: What are the most common reasons my synthetic gene circuit loses function over time? Circuit failure often stems from genetic instability. The primary cause is the metabolic burden imposed by the circuit, which slows host cell growth. This creates a selective pressure for faster-growing mutants that have inactivated or lost the circuit function. Common failure modes include plasmid loss due to segregation errors, recombination-mediated deletion of repeated genetic sequences, and disruptive insertions from transposable elements [21].
FAQ 2: How can I improve the long-term evolutionary stability of my recombinase-based memory device? Two complementary strategies are "suppressing mutant emergence" and "suppressing the relative fitness of mutants." You can achieve this by:
- Genomic Integration: Integrating your circuit into the host genome to prevent plasmid loss [21].
- Reducing Genetic Repeats: Minimizing the use of repeated sequences (like identical promoters or terminators) to lower the risk of recombination [21].
- Using Reduced-Genome Hosts: Employing engineered chassis organisms with deleted transposable elements to lower the background mutation rate [21].
- Implementing Genetic Controllers: Designing feedback controllers that sense circuit output or host growth rate and adjust expression to reduce burden, thereby extending functional half-life [2].
FAQ 3: Why does my circuit function correctly in a test tube but fail inside a mammalian cell? This is frequently due to context dependence and host-circuit interactions. A circuit that is well-characterized in one organism (e.g., E. coli) may behave unpredictably in another (e.g., mammalian cells) due to differences in endogenous machinery, resource pools, and unintended interactions with native cellular components [22]. Furthermore, the metabolic burden of the circuit can differ significantly between hosts, leading to toxic effects or strong selection against circuit-bearing cells [22] [21].
FAQ 4: What can I do if my circuit shows high cell-to-cell variability (noise) in its output? High noise often results from the stochastic nature of biochemical reactions involving small numbers of molecules. This can be intrinsic (from the circuit itself) or extrinsic (from global fluctuations in cellular resources). Strategies to address this include:
- Using Orthogonal Parts: Employing genetic components (e.g., polymerases, ribosomes) that do not interact with the host's native systems to decouple from global fluctuations [22].
- Implementing Feedback Control: Negative feedback architectures can suppress variation and stabilize output [2].
- Characterizing Parts in Context: Using cell-free systems for rapid, high-throughput characterization of genetic parts before implementing them in living cells [22].
Troubleshooting Guides
Problem: Low or No Circuit Output
| Potential Cause | Diagnostic Experiments | Recommended Solutions |
|---|---|---|
| Part Failure | Sequence circuit to check for mutations; Verify part activity with a reporter assay in a validated host. | Re-clone the defective part; Use well-characterized, high-quality parts from repositories. |
| Host-Circuit Incompatibility | Measure host growth rate with and without circuit; Perform RNA-seq to identify unintended interactions [22]. | Switch to a more compatible chassis (e.g., reduced-genome strain); Refactor the circuit to use more orthogonal parts [21]. |
| Excessive Metabolic Burden | Quantify the reduction in host growth rate upon circuit activation [21]. | Lower constitutive expression levels; Use inducible systems; Implement burden-aware feedback controllers [2]. |
| Incorrect Assembly (Recombinase Circuits) | Verify the orientation of genetic elements (promoters, genes) flanked by recombinase sites after induction [23] [24]. | Ensure the DNA sequence is "well-formed" per syntactic rules; Confirm recombinase specificity and activity [23]. |
Problem: Genetic Instability and Loss of Memory
| Potential Cause | Diagnostic Experiments | Recommended Solutions |
|---|---|---|
| Plasmid Loss | Plate cells on selective and non-selective media to count plasmid-retaining colonies. | Integrate the circuit into the host chromosome; Use stable, low-copy-number plasmids [21]. |
| Evolutionary Escape | Serial passage cells for multiple generations and track functional output and population genetics [21] [2]. | Couple circuit function to an essential gene (e.g., for antibiotic resistance); Use kill-switch circuits to eliminate non-functional cells [21]. |
| Mutation in Key Parts | Isolate non-functional cells and sequence the entire circuit to identify inactivating mutations. | Avoid repeated sequences; Use robust, host-aware genetic designs; Distribute large populations into smaller, segregated compartments to confine mutants [21]. |
Problem: Unintended Logic or Leaky Expression
| Potential Cause | Diagnostic Experiments | Recommended Solutions |
|---|---|---|
| Recombinase Crosstalk | Test each recombinase individually and in combination for orthogonality. | Use bioinformatic tools to select highly orthogonal recombinase/att-site pairs [24]. |
| Promoter Interference | Characterize the activity of each promoter in isolation and in the final circuit context [22]. | Re-design circuit layout; Introduce insulating sequences between genetic parts. |
| Resource Overload | Model resource allocation (ribosomes, nucleotides) to identify potential bottlenecks [22]. | Re-balance expression levels of circuit components; Use resource-aware whole-cell models during the design phase [25]. |
Experimental Protocols for Key Experiments
Protocol 1: Serial Passaging to Quantify Evolutionary Stability
Purpose: To experimentally determine the functional half-life of a synthetic gene circuit under prolonged cultivation.
Materials:
- Engineered bacterial strain with gene circuit.
- Appropriate liquid growth medium (e.g., LB).
- Sterile flasks or culture tubes.
- Incubator/shaker.
- Flow cytometer or plate reader for output measurement (e.g., GFP).
Methodology:
- Inoculation: Start a batch culture by inoculating the engineered strain into fresh medium.
- Growth and Dilution: Grow the culture for a set period (e.g., 24 hours). Each day, dilute an aliquot of the culture into fresh medium to maintain continuous growth. A typical dilution is 1:100 to 1:1000.
- Monitoring: At each passage, sample the culture to:
- Measure Circuit Output: Use fluorescence or other assays to quantify the population-level function.
- Measure Growth Rate: Track optical density (OD) over time.
- Check for Mutants: Plate cells to isolate single colonies and screen for loss-of-function phenotypes.
- Analysis: Continue for 10+ generations. Plot the population-level output over time. The "half-life" (τ50) is the time taken for the output to fall to 50% of its initial value [2].
Protocol 2: Validating Recombinase-Based Logic Gates
Purpose: To confirm the correct truth table operation of a recombinase-based logic gate.
Materials:
- Bacterial strains harboring the recombinase-based logic circuit.
- Chemical inducers for the specific recombinase promoters (e.g., AHL, aTc).
- Solid and liquid growth media.
- Flow cytometer or fluorescence microscope.
Methodology:
- Induction: For each possible input combination (e.g., No inducer, Inducer A, Inducer B, Both inducers), grow separate cultures and add the corresponding inducers.
- Incubation: Allow sufficient time for recombinase expression and DNA recombination to occur (typically 12-24 hours).
- Measurement: Analyze the output (e.g., GFP fluorescence) using flow cytometry to determine the ON/OFF state for each input condition.
- Verification: Isolate genomic DNA from cells in each state and perform PCR or sequencing across the recombinase sites to confirm the expected DNA inversion or excision pattern matches the logical output [23] [24].
The Scientist's Toolkit: Research Reagent Solutions
| Item | Function & Application |
|---|---|
| Orthogonal Serine Recombinases (Bxb1, phiC31) | Enzyme that catalyzes irreversible DNA inversion, excision, or integration between specific attP and attB sites. Used as the core processor in logic and memory circuits [23] [24]. |
| Well-Formed Sequence (WFS) DNA Constructs | A syntactically correct DNA sequence where genetic elements (promoters, genes) are flanked by recombinase targeting sites. Essential for predictable circuit behavior [23]. |
| Reduced-Genome E. coli Strains (e.g., MDS42) | Chassis with transposable elements and genomic islands removed. Reduces the rate of insertion-sequence-mediated circuit failure, enhancing genetic stability [21]. |
| Host-Aware Model Framework | A computational model that simulates interactions between circuit expression and host resource pools (ribosomes, energy). Used to predict burden and evolutionary dynamics in silico before building the circuit [2]. |
| Genetic Feedback Controllers | A synthetic module that senses a signal (e.g., circuit output, growth rate) and uses negative feedback to actuate the circuit (e.g., via sRNAs). Mitigates burden and extends functional longevity [2]. |
Signaling Pathway and Workflow Diagrams
Diagram: Recombinase Logic Gate Mechanism
Diagram: Circuit Stability Troubleshooting Workflow
FAQs: Core Concepts and Applications
FAQ 1: What is the primary functional advantage of using transcriptional condensates to stabilize synthetic gene circuits?
The primary advantage is the ability to buffer against growth-mediated dilution. As cells grow and divide, key transcription factors (TFs) in synthetic circuits become diluted, leading to circuit failure. Transcriptional condensates concentrate these TFs at their target promoters, creating a local high-concentration environment that is resilient to global dilution in the cell, thereby maintaining consistent gene expression and circuit function across cell generations [8] [15].
FAQ 2: My synthetic circuit loses its bistable memory after cell division. Can phase separation help?
Yes. This is a common failure mode where dilution of TFs disrupts the self-reinforcing loop of a bistable switch. By fusing an Intrinsically Disordered Region (IDR) to your circuit's transcription factor, you can promote the formation of condensates. These condensates maintain a high local TF concentration at the promoter, which preserves the bistable "ON" state even after rapid cell growth and division, effectively restoring the circuit's memory function [15].
FAQ 3: Are transcriptional condensates a form of irreversible, hard-wired memory for cells?
No. Condensates stabilized through phase separation provide a form of dynamic and reversible memory. Unlike memory circuits built with DNA recombinases that create permanent, irreversible genetic changes [10], condensate-based stabilization is physical and tunable. The condensates can form and dissolve in response to cellular conditions, allowing for dynamic control while still providing protection against dilution during growth phases [8] [26].
FAQ 4: What is the most critical component for engineering phase separation into a synthetic circuit?
The most critical components are Intrinsically Disordered Regions (IDRs). IDRs are protein domains that lack a fixed 3D structure and facilitate weak, multivalent interactions. By fusing a well-characterized IDR (e.g., from the FUS protein or a synthetic resilin-like polypeptide) to your transcription factor, you can drive the assembly of biomolecular condensates at the promoter site [8] [15].
Troubleshooting Guides
Guide 1: My Condensates Are Not Forming
| Symptom | Possible Cause | Solution |
|---|---|---|
| No visible condensates under microscope. | The fused IDR has weak phase-separation capability. | Switch to a stronger IDR, such as the N-terminal domain of FUS (FUSn) or a synthetic RLP [15]. |
| Diffuse fluorescence throughout the cell. | The expression level of the fusion protein is too low to reach the concentration threshold for phase separation. | Optimize the promoter strength or ribosome binding site (RBS) to increase protein expression [15]. |
| Condensates form in the wrong cellular location. | The fusion protein lacks proper localization signals. | Include localization sequences in your construct to target the transcription factor and its condensate to the nuclear or promoter region. |
Guide 2: My Circuit Memory is Still Unstable
| Symptom | Possible Cause | Solution |
|---|---|---|
| Circuit loses memory after prolonged growth. | Condensates are dissipating or not dense enough to maintain a critical TF concentration. | Experiment with different IDRs to tune the stability and physical properties of the condensates. Validate via FRAP to ensure they are liquid-like and dynamic [15]. |
| High cell-to-cell variability in memory retention. | Stochastic formation or dissolution of condensates in individual cells. | Use a stronger, more consistent promoter to express the TF-IDR fusion protein and reduce expression noise. Model the system to understand the stochastic dynamics [15]. |
| The circuit imposes a high metabolic burden. | Overexpression of synthetic genes drains cellular resources. | Consider integrating a resource allocation controller or optimizing codon usage to reduce the burden on the host cell [15]. |
Experimental Protocols
Protocol 1: Constructing a Phase-Separation-Stabilized Gene Circuit
This protocol outlines the key steps for engineering a synthetic self-activation (SA) circuit that uses transcriptional condensates for stability, based on the Droplet-Self-Activation (Drop-SA) design [15].
1. Design and Cloning:
- Core Circuit: Assemble a bicistronic self-activation circuit where the transcription factor (e.g., AraC) and a reporter (e.g., GFP) are under the control of a inducible promoter (e.g., Pbad).
- IDR Fusion: Genetically fuse a selected IDR (e.g., FUSn or RLP20) to the C-terminus of the transcription factor. For visualization and validation, the construct can be a triple fusion: GFP-IDR-TF (e.g., GFP-FUSn-AraC).
- Cloning: Use standard molecular biology techniques (e.g., Gibson assembly, Golden Gate) to clone the final construct into an appropriate plasmid vector.
2. Transformation and Cell Culture:
- Transform the constructed plasmid into your host cell line (e.g., E. coli).
- Culture the cells in a suitable medium. To test the circuit, grow cells to mid-log phase and activate the circuit by adding an inducer (e.g., L-arabinose for the Pbad promoter).
3. Validation and Imaging:
- Condensate Formation: Use fluorescence microscopy to check for the formation of bright, punctate droplets (condensates) within the cells, typically localizing at the cell poles in bacteria.
- Liquid Property Validation: Perform Fluorescence Recovery After Photobleaching (FRAP) on the droplets. Photobleach a region of interest and monitor the recovery of fluorescence over time. Rapid recovery (e.g., within 10-15 minutes) confirms the liquid-like nature of the condensates and dynamic exchange of molecules [15].
Protocol 2: Validating Circuit Performance with FRAP
1. Sample Preparation:
- Prepare cells expressing your Drop-SA circuit and mount them on an agarose pad for microscopy.
2. Photobleaching and Recovery:
- Pre-bleach: Capture an image of the condensate.
- Bleach: Use a high-intensity laser to bleach the fluorescence in a defined region of the condensate.
- Recovery: Immediately after bleaching, capture images at regular time intervals (e.g., every 10 seconds for 15 minutes) to monitor the fluorescence signal returning to the bleached area.
3. Data Analysis:
- Quantify the fluorescence intensity in the bleached area over time.
- Normalize the intensities to the pre-bleach and post-bleach levels.
- Plot the recovery curve. A hallmark of liquid-liquid phase separation is a rapid, significant recovery of fluorescence, indicating high dynamics within the condensate [15].
Data Presentation
Table 1: Comparison of Intrinsically Disordered Regions (IDRs) for Engineering Condensates
| IDR Name | Origin | Key Features | Phase Separation Behavior | Example Use in Circuits |
|---|---|---|---|---|
| FUSn | Human FUS protein | Well-characterized, natural IDR; promotes strong condensate formation [15]. | UCST-type (forms at lower temperatures) [15]. | Fused to AraC in a self-activation circuit to restore bistable memory [15]. |
| RLP20 | Synthetic resilin-like polypeptide | Engineered polypeptide; properties can be tuned [15]. | UCST-type (forms at lower temperatures) [15]. | Fused to AraC to form condensates and buffer against dilution [15]. |
Table 2: Quantitative Analysis of Circuit Performance With and Without Phase Separation
| Circuit Type | Bistable Memory Retention (After Dilution) | Hysteresis Range (Under Growth Dilution) | Key Experimental Evidence |
|---|---|---|---|
| Standard Self-Activation (SA) Circuit | Rapid memory loss; transitions to OFF state [15]. | Loss of hysteresis; dose-response becomes independent of initial condition [15]. | Dilution of TF concentration leads to circuit failure [15]. |
| Droplet-SA (Drop-SA) Circuit | Robust memory retention; recovers to ON state [15]. | Hysteresis range restored to near pre-dilution levels [15]. | FRAP confirms liquid condensates; local TF concentration at promoter is maintained [15]. |
Visualization Diagrams
Diagram 1: Stabilizing a Gene Circuit via Transcriptional Condensates
Diagram 2: Experimental Workflow for Building a Drop-SA Circuit
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for Engineering Transcriptional Condensates
| Reagent | Function | Example & Notes |
|---|---|---|
| Intrinsically Disordered Regions (IDRs) | Drives phase separation via multivalent, weak interactions [8] [15]. | FUSn (1-267): A well-established natural IDR [15]. RLP20: A synthetic, tunable resilin-like polypeptide [15]. |
| Fluorescent Reporter Proteins | Allows visualization of condensate formation and circuit output. | Green Fluorescent Protein (GFP): Often fused to the TF-IDR construct for direct visualization of the condensates [15]. |
| Inducible Promoter Systems | Provides external control over the expression of the synthetic circuit. | Pbad (Arabinose-inducible): Used to activate the circuit in the cited studies [15]. Other systems (Tet-On, GAL) are also applicable. |
| Cre Recombinase (or other recombinases) | Useful for creating irreversible memory circuits or, in other systems, for editing genetic set points as a comparison/alternative strategy [10] [3]. | Bacteriophage integrases (PhiC31, Bxb1) or yeast recombinases (Flp) can be used to build complex logic [10]. |
Practical Strategies for Diagnosing and Fixing Circuit Instability
Frequently Asked Questions
What is metabolic burden, and why is it a problem for my gene circuit? Metabolic burden is the fitness cost imposed on a host cell by an engineered gene circuit. It occurs because the circuit consumes limited cellular resources—such as ribosomes, energy (anabolites), and nucleotides—for its own expression, diverting them from the host's natural processes [2]. This often reduces cell growth rate. Since growth rate is directly tied to fitness in microbes, cells with functioning circuits are outcompeted by faster-growing, non-producing mutant cells that inevitably arise in the population, leading to a rapid loss of circuit function [2].
My circuit's protein output is inconsistent across a population of cells. How can I fix this? Inconsistent output often stems from variations in gene copy number between cells and natural biological noise. A solution is to implement a control system like the DIAL (Dialable Expression) system. This system uses a spacer sequence between the promoter and the gene of interest; a longer spacer results in lower expression. By incorporating sites for recombinase enzymes (e.g., Cre recombinase) into this spacer, you can precisely edit the expression "set point" after delivery, enabling uniform protein levels across a cell population [3].
The function of my circuit degrades over multiple cell generations. What strategies can improve its evolutionary longevity? Evolutionary degradation occurs due to mutations that reduce circuit burden. Implementing genetic feedback controllers can significantly extend functional half-life [2]. Key strategies include:
- Negative Autoregulation: Prolongs short-term performance [2].
- Growth-Based Feedback: Extends the functional half-life of the circuit [2].
- Post-Transcriptional Control: Using small RNAs (sRNAs) for silencing often outperforms transcriptional control by enabling strong regulation with lower burden [2].
- Transcriptional Condensates: Forming droplet-like compartments around key genes via liquid-liquid phase separation can shield synthetic programs from being diluted during cell division, maintaining stability across generations [27].
What are the key metrics for quantifying burden and evolutionary stability? When simulating or experimenting with an evolving population, you can use the following key metrics [2]:
- P₀: The initial total protein output of the ancestral population before any mutation.
- τ±₁₀: The time taken for the total output (P) to fall outside the range of P₀ ± 10%. This measures short-term stability.
- τ₅₀: The time taken for the total output to fall below P₀/2. This measures long-term "persistence."
Troubleshooting Guides
Problem: Rapid Loss of Circuit Function in Serial Passaging
| Symptoms | Possible Causes | Diagnostic Experiments | Solutions |
|---|---|---|---|
| Fluorescence or protein output declines rapidly over 24-72 hours [2]. | High metabolic burden enriching for loss-of-function mutants [2]. | Sequence circuit elements from the population at different time points to identify common mutations [2]. | Implement growth-based feedback controllers [2]. |
| Culture growth rate increases as circuit function decreases [2]. | Mutation in promoters, RBS, or coding sequences that reduce resource consumption [2]. | Measure growth rates of isolated clones; mutants often grow faster [2]. | Use post-transcriptional control (sRNAs) for more efficient regulation [2]. |
| Mutations are inevitable in large populations; selection is the primary driver [2]. | Flow cytometry to analyze population heterogeneity in protein expression. | Couple circuit function to an essential gene (e.g., antibiotic resistance) [2]. |
Problem: High Cell-to-Cell Variability in Protein Expression
| Symptoms | Possible Causes | Diagnostic Experiments | Solutions |
|---|---|---|---|
| Broad, non-bimodal distribution of fluorescence in flow cytometry [3]. | Variation in the number of circuit copies taken up by individual cells [3]. | Flow cytometry to quantify the distribution of expression levels. | Implement the DIAL system to edit expression set points post-delivery for uniformity [3]. |
| Unstable cell fate reprogramming (e.g., in fibroblast-to-neuron conversion) [3]. | Natural biological variation in transcription factor levels and resource availability [3]. | Single-cell RNA sequencing to correlate transcript levels with phenotype. | Use a system like ComMAND in conjunction with DIAL to prevent overexpression and maintain consistent levels [3]. |
Quantitative Metrics for Burden and Evolutionary Longevity
The table below summarizes key metrics and methods used to assess the impact of synthetic gene circuits on host cells.
| Metric | Description | Measurement Technique | Interpretation |
|---|---|---|---|
| Initial Output (P₀) | Total target protein molecules produced by the entire ancestral population before mutation [2]. | Measure total fluorescence (e.g., GFP) or protein concentration via spectrophotometry or ELISA at time zero. | Higher P₀ indicates strong initial circuit function, but often correlates with higher burden [2]. |
| Stability Time (τ±₁₀) | Time for population-wide protein output to fall outside P₀ ± 10% [2]. | Time-course tracking of output (e.g., fluorescence) in batch culture or serial passaging. | A longer τ±₁₀ indicates better short-term performance maintenance [2]. |
| Functional Half-Life (τ₅₀) | Time for population-wide protein output to fall below 50% of P₀ [2]. | Time-course tracking of output during long-term culture. | A longer τ₅₀ indicates greater evolutionary longevity and persistence of some circuit function [2]. |
| Relative Growth Rate | Growth rate of circuit-carrying cells relative to unengineered cells [2]. | Measure optical density (OD600) or cell counts over time in parallel cultures. | A ratio < 1 indicates significant metabolic burden. |
Experimental Protocols
Protocol 1: Measuring Evolutionary Longevity in Serial Passaging
Objective: To determine the functional half-life (τ₅₀) of a synthetic gene circuit in a microbial population.
- Inoculation: Start a batch culture from a single colony of the engineered strain.
- Sampling and Measurement: At a defined time point each day (e.g., 24 hours): a. Measure Output: Take a sample and measure the population-level output (e.g., fluorescence/OD600 for reporters). b. Passage: Dilute the culture into fresh medium to reset the population density and nutrient levels, mimicking repeated batch conditions [2].
- Data Analysis: Plot the total output (P) over time. Calculate τ±₁₀ and τ₅₀ based on the initial output P₀ [2].
Protocol 2: Implementing the DIAL System for Set-Point Control
Objective: To achieve uniform and adjustable gene expression levels after circuit delivery.
- Circuit Design: Clone your gene of interest downstream of a promoter, with a long, engineered DNA spacer placed between them. Incorporate multiple recombinase excision sites (e.g., lox sites for Cre recombinase) within this spacer [3].
- Delivery: Transfer the constructed DIAL circuit into the target cells using an appropriate method (e.g., viral transduction, electroporation).
- Set-Point Tuning: At any point after delivery, transfert the cells with a plasmid expressing Cre recombinase (or other specific recombinases). The level and combination of recombinases will determine the final length of the spacer, thereby setting the expression level to "high," "med," or "low" [3].
- Validation: Use flow cytometry 48-72 hours post-recombinase delivery to confirm uniform protein expression across the cell population.
The Scientist's Toolkit: Research Reagent Solutions
| Reagent / Tool | Function in Quantifying/Reducing Burden |
|---|---|
| DIAL System | A genetic device that allows post-delivery, fine-tuned adjustment of gene expression levels to ensure uniformity and reduce burden from overexpression [3]. |
| Cre Recombinase | An enzyme used in the DIAL system to precisely excise parts of the DNA spacer, bringing the promoter closer to the gene and dialing up expression [3]. |
| Host-Aware Model | A multi-scale computational framework that simulates host-circuit interactions, mutation, and population dynamics to predict evolutionary longevity and guide controller design [2]. |
| Genetic Controllers | Feedback architectures (e.g., using sRNAs or transcription factors) that sense circuit output or growth rate and adjust expression to maintain stability and reduce burden [2]. |
| Transcriptional Condensates | Engineered droplet-like compartments formed via liquid-liquid phase separation that protect synthetic genes from dilution during cell division, enhancing long-term stability [27]. |
Signaling Pathways and Workflows
Welcome to the Technical Support Center
This resource is designed to help researchers troubleshoot common issues related to the evolutionary stability of synthetic gene circuits. Below you will find FAQs, troubleshooting guides, and detailed experimental protocols focused on maintaining circuit function against mutational failure.
Frequently Asked Questions
FAQ 1: Why does my synthetic gene circuit lose function within a few generations of microbial growth?
Circuit failure primarily occurs due to mutations that reduce the metabolic burden imposed on the host cell [2] [21]. Engineered circuits consume cellular resources like nucleotides, amino acids, and ribosomes, diverting them from host processes essential for growth and replication. This "burden" creates a selective pressure where any cell with a mutation that inactivates or reduces circuit function will grow faster and outcompete the ancestral, circuit-carrying strain [2]. Mutations can occur in circuit elements like promoters, ribosome binding sites, or coding sequences [21].
FAQ 2: What are the key metrics for quantifying the evolutionary longevity of a gene circuit?
The field has coalesced around several key metrics to standardize the measurement of evolutionary longevity [2]:
- Initial Output (P₀): The total functional output of the circuit in the ancestral population before any mutations arise.
- Functional Maintenance Time (τ±₁₀): The time taken for the population-level circuit output to fall outside a 10% window of P₀.
- Functional Half-Life (τ₅₀): The time taken for the population-level circuit output to fall below 50% of P₀. This measures the long-term "persistence" of circuit function.
The table below summarizes a quantitative comparison of different controller architectures from a recent in silico study [2].
Table 1: Performance Metrics of Different Genetic Controller Architectures
| Controller Architecture | Key Mechanism | Impact on Short-Term Performance (τ±₁₀) | Impact on Long-Term Half-Life (τ₅₀) | Relative Burden |
|---|---|---|---|---|
| Open-Loop (No Control) | N/A | Baseline | Baseline | High |
| Negative Autoregulation | Transcriptional feedback on circuit's own gene | Prolonged | Moderate improvement | Reduced |
| Growth-Based Feedback | Actuation tied to host growth rate | Moderate improvement | Significantly extended (superior long-term) | Low |
| Post-Transcriptional Control | Feedback via small RNAs (sRNAs) | Good improvement | Good improvement | Very Low |
| Multi-Input Controllers | Combines e.g., growth and output sensing | Optimized | Over 3x improvement vs. open-loop | Tunable |
FAQ 3: My circuit is stable in lab-scale cultures but fails in industrial fermenters. Why?
This is a classic problem of population size-dependent mutant emergence [21]. The probability of a mutant appearing in a population is directly proportional to the number of cell divisions. A large-scale fermenter provides a vastly larger population and more generations than a lab-scale culture, dramatically increasing the chance that a loss-of-function mutant will emerge and take over the population [21]. Strategies like metabolic engineering to reduce burden or using genomic integration instead of plasmids are crucial for scaling up.
FAQ 4: Are there ways to design circuits that are inherently more robust to evolution?
Yes. Beyond reducing burden, two main engineering strategies are employed [21]:
- Suppress Mutant Emergence: This involves reducing the rate at which mutants appear. Methods include using engineered host strains with reduced mutation rates (e.g., by removing transposable elements), minimizing repetitive DNA sequences in the circuit to avoid recombination, and using genomic integration instead of error-prone plasmids [21].
- Suppress Mutant Fitness: This involves making the circuit function essential for survival, so that mutants lose their fitness advantage. This can be done by coupling the output to the expression of an essential gene or using toxin-antitoxin systems [28].
Troubleshooting Guides
Problem: Rapid Dominance of Non-Functional Mutants
Symptoms: A rapid decline in population-average circuit output (e.g., fluorescence) over serial passages, accompanied by a measurable increase in population growth rate.
Solutions:
- Implement a Genetic Feedback Controller.
- Rationale: Negative feedback can buffer the system against fluctuations and reduce the resource burden, thereby lowering the selective advantage of non-producers [2].
- Design Choice: Post-transcriptional controllers using small RNAs (sRNAs) often outperform transcriptional ones because they provide strong control with lower burden [2].
- Implementation: See Experimental Protocol 1 below.
Couple Circuit Function to Cell Survival.
- Rationale: If the circuit is essential for survival under your culture conditions, mutants that lose the circuit will die [21].
- Method: Use a bidirectional promoter that simultaneously drives your gene of interest and an antibiotic resistance gene. In the presence of the antibiotic, only cells retaining the functional circuit will survive [28]. Note: Mutations can still occur in the RBS, so this is not foolproof [2].
Utilize a "Host-Aware" Framework.
- Rationale: Model the circuit and its interaction with the host (resource competition, growth impact) in silico before building it. This allows you to predict evolutionary dynamics and optimize controller parameters for stability [2].
Problem: High Leaky Expression in Feedback Circuits
Symptoms: Unacceptably high basal output even in the "off" state, which can impose a constant burden and accelerate evolution.
Solutions:
- Fine-Tune Controller Set Points with a System like DIAL.
- Rationale: Leakiness can be caused by suboptimal spacing between genetic parts. The DIAL system allows you to edit the expression level after circuit delivery [3].
- Implementation: Incorporate a spacer sequence between the promoter and the gene of interest. By using recombinase enzymes (e.g., Cre) to excise parts of this spacer, you can adjust the distance and fine-tune the expression level to find an optimal balance between output and burden [3].
- Adopt a Multi-Input Controller Architecture.
- Rationale: Leaky expression from one sensor can be gated by a second input. This is the principle of an AND gate.
- Implementation: Design a circuit that requires two inputs (e.g., a specific metabolite AND a specific temperature) to activate the output. This drastically reduces the chance of spurious activation and the resulting burden [29] [4].
Experimental Protocols
Protocol 1: Implementing a Post-Transcriptional Feedback Controller for Enhanced Longevity
Principle: Use a small RNA (sRNA) to silence the mRNA of your circuit's gene, creating a negative feedback loop that reduces burden and stabilizes output [2].
Materials:
- Plasmid Backbone: Standard cloning vector for your host (e.g., high-copy for E. coli).
- Target Gene: Your gene of interest (GOI), e.g., a fluorescent reporter.
- sRNA Scaffold: A plasmid containing a constitutive promoter driving an sRNA sequence complementary to the 5' UTR of your GOI's mRNA.
- Host Strain: Appropriate competent cells (e.g., E. coli DH10B).
Workflow:
- Circuit Construction: Clone your GOI into the plasmid under a constitutive promoter.
- Sensor Module: Design an sRNA sequence that is antisense to the ribosome binding site (RBS) and/or the start codon of your GOI's mRNA. Clone this sRNA into a compatible plasmid under a constitutive promoter.
- Co-transformation: Transform both plasmids into your host strain.
- Validation:
- Measure the expression level of your GOI and the growth rate of the controlled strain versus an open-loop control (GOI without the sRNA controller).
- You should observe a reduction in both expression level and burden (higher growth rate) in the controlled strain.
- Evolutionary Experiment:
- Serially passage both the controlled and open-loop strains for ~50-100 generations.
- Sample the population periodically to measure the population-average output (P) and the fraction of cells still expressing the GOI (e.g., via flow cytometry).
- Calculate the functional half-life (τ₅₀) for both populations.
The following diagram illustrates the logical workflow of this experiment.
Protocol 2: Testing Circuit Stability Using Serial Passage
Principle: Simulate long-term evolution in a laboratory setting to directly measure the evolutionary half-life (τ₅₀) of your circuit [2].
Materials:
- Strains: Your engineered strain and a control (e.g., unengineered wild-type).
- Media: Appropriate liquid growth medium (e.g., LB for E. coli).
- Equipment: shaking incubator, spectrophotometer, plate reader or flow cytometer.
Workflow:
- Inoculation: Start a primary culture from a single colony and grow to mid-exponential phase.
- Dilution and Passage: Every 24 hours, dilute the culture into fresh medium. A standard dilution is 1:1000, which allows for about 10 generations per day.
- Monitoring:
- Growth: Measure the optical density (OD) at the beginning and end of each 24-hour cycle to track fitness.
- Function: Sample the population at each passage and measure the circuit output (e.g., fluorescence). For a more detailed view, use flow cytometry to track the distribution of expression across the population.
- Data Analysis:
- Plot the total population output (P) over time.
- Fit a curve to the data and calculate the time taken for the output to drop to half of its initial value (τ₅₀).
The Scientist's Toolkit
Table 2: Research Reagent Solutions for Circuit Longevity Research
| Reagent / Tool | Function in Experiment | Key Consideration |
|---|---|---|
Reduced-Genome Host Strains (e.g., MDS42) |
Engineered hosts with transposable elements removed to lower background mutation rates [21]. | Reduces mutation-driven failure, simplifying the analysis of circuit-specific evolutionary pressures. |
| Orthogonal Parts (e.g., Bacterial TFs in Plants) | Genetic parts that function independently of the host's native networks [4]. | Minimizes host-circuit interference (cross-talk), improving predictability and reducing unintended fitness costs. |
| Genomic Integration Tools (e.g., CRISPR) | Stably inserts the circuit into the host chromosome [21]. | Avoids plasmid loss from segregation errors; typically offers lower, more stable copy number than plasmids. |
| Phase-Separation Inducing Tags (e.g., IDR Fusions) | Fuses Intrinsically Disordered Regions (IDRs) to transcription factors to form condensates [8]. | Creates membraneless organelles that concentrate circuit components, buffering against growth-mediated dilution. |
| DIAL System Components | Set of recombinase target sites for post-hoc tuning of promoter-gene distance [3]. | Allows for fine-tuning of expression levels after circuit construction to find a burden-stability optimum. |
| High-Throughput Evolution Platforms (e.g., eVOLVER) | Automated systems for running parallel, controlled evolution experiments [11]. | Accelerates the testing of circuit longevity under various selective pressures by scaling up experimental throughput. |
Pathway Diagram: Multi-Input Controller for Robust Cancer Targeting
The following diagram illustrates a sophisticated AND-gate circuit design, which achieves high selectivity in therapeutic applications by requiring multiple cancer-specific signals for activation [29].
This technical support center provides troubleshooting guides and FAQs for researchers facing challenges with synthetic gene circuit expression and stability. The resources below address common issues by applying orthogonal design principles to minimize interference with host cellular processes.
Troubleshooting Guides
Problem 1: Rapid Loss of Circuit Function Due to Evolution
Problem Description: The synthetic gene circuit loses function over multiple cell generations. The population-level output of your protein of interest declines significantly during prolonged culture or fermentation.
Diagnosis Questions:
- Has the growth rate of your engineered cell population increased over time?
- Are you observing genetic mutations in the circuit's regulatory elements (e.g., promoters)?
- Does the problem occur more rapidly with circuits that express genes at high levels?
Solution: Implement genetic feedback controllers to stabilize expression and reduce the selective advantage of mutant cells.
- Underlying Cause: Synthetic circuits consume cellular resources (ribosomes, nucleotides, amino acids), imposing a metabolic burden that reduces host cell growth rate. Cells with mutated, non-functional circuits grow faster and outcompete the engineered cells [2].
- Corrective Action: Introduce a control system that dynamically regulates circuit expression to minimize burden.
Experimental Protocol: Implementing a Negative Feedback Controller This protocol outlines the creation of a transcriptional negative feedback loop to stabilize expression [2].
- Circuit Design: Clone your gene of interest (GOI) downstream of a promoter that is repressed by a transcription factor (TF).
- Controller Integration: Design a constitutive promoter to express the repressor TF. Also, express the same TF under the control of a promoter that is activated by the GOI's protein product (or a direct reporter like GFP). This creates the feedback loop.
- Assembly and Transformation: Assemble the complete circuit and transform it into your host organism (e.g., E. coli).
- Validation Experiments:
- Conduct a long-term growth experiment, serially passaging the culture for multiple generations.
- Measure both the population-level output (e.g., total fluorescence) and host cell growth rate at regular intervals.
- Compare the performance against an open-loop control (the same circuit without the feedback mechanism).
Performance Comparison of Control Strategies [2]
| Control Strategy | Mechanism | Short-Term Stability (τ±10) | Long-Term Half-Life (τ50) | Key Advantage |
|---|---|---|---|---|
| Open-Loop (No Control) | Constitutive expression | Low | Low | Simple design, high initial output |
| Transcriptional Feedback | TF represses GOI transcription | Medium | Medium | Good burden reduction |
| Post-Transcriptional Feedback | sRNA silences GOI mRNA | High | High | Stronger control, lower controller burden |
| Growth-Based Feedback | Circuit expression linked to host growth rate | Low | Very High | Maximizes long-term evolutionary persistence |
Problem 2: Heterogeneous and Uncontrolled Gene Expression
Problem Description: There is significant cell-to-cell variability in protein expression levels from your synthetic circuit. You cannot reliably set or adjust the expression level after delivery.
Diagnosis Questions:
- Does your cell population show a wide distribution of output intensity when measured by flow cytometry?
- Is it difficult to achieve a specific, uniform threshold of protein expression across the entire cell population?
- Do you need the ability to tune the expression level post-transduction?
Solution: Use a modular, distance-based system to precisely dial expression levels.
- Underlying Cause: Natural variation in cellular components and random differences in plasmid copy number upon delivery lead to inconsistent expression [3].
- Corrective Action: Implement a system where the physical distance between a promoter and a gene acts as a tunable set point.
Experimental Protocol: Tuning with the DIAL System This protocol uses the DIAL (Distance-Induced Adjustment of Levels) system, which employs Cre recombinase to edit spacer sequences [3].
- Circuit Construction: Assemble your transgene with a promoter separated by a long DNA "spacer" region. This spacer should contain multiple loxP sites (or sites for other recombinases).
- Delivery: Transfect the circuit into your target cells (e.g., mammalian cells).
- Expression Tuning:
- Low/Off: No Cre is added. The long spacer keeps expression low.
- Medium: Add a low concentration of Cre to excise part of the spacer.
- High: Add a high concentration of Cre to excise most of the spacer, bringing the promoter close to the gene.
- Validation: Use flow cytometry 48-72 hours after Cre delivery to quantify the distribution and mean of protein expression in the population.
Problem 3: High Metabolic Burden and Resource Competition
Problem Description: The host cells exhibit severely slowed growth after circuit introduction. The circuit's function may also be impaired due to competition for limited transcriptional/translational resources.
Diagnosis Questions:
- Does the doubling time of your engineered strain significantly exceed that of the wild-type?
- Does high-level expression of your circuit lead to a complete halt in cell growth?
- Does the circuit performance degrade when multiple genes are expressed simultaneously?
Solution: Decouple the circuit from host processes using orthogonal components and physical compartmentalization.
- Underlying Cause: The host's native gene expression machinery (RNA polymerases, ribosomes) is hijacked by the synthetic circuit, depleting resources for essential host functions [30] [2].
- Corrective Action: Employ orthogonal design to create independent functional modules.
Experimental Protocol: Leveraging Phase Separation for Stability This protocol describes using liquid-liquid phase separation to create transcriptional condensates that shield synthetic circuits [27].
- Identify Condensate-Forming Proteins: Select proteins or RNA molecules known to undergo phase separation (e.g., certain transcription factors or disordered proteins).
- Fusion Protein Design: Fuse these proteins to components of your synthetic gene circuit (e.g., to a transcription factor or activator).
- Circuit Assembly and Testing: Integrate these engineered components into your circuit and deliver it to cells.
- Imaging and Validation:
- Use live-cell microscopy to confirm the formation of condensate droplets around the circuit's genes.
- Compare the growth rate and long-term functional stability of cells with and without the engineered condensates over multiple generations.
Frequently Asked Questions (FAQs)
Q: What does "orthogonal" mean in the context of synthetic biology? A: Orthogonal design means creating system components (like genetic parts) that function independently from the host's native processes. A change in one should not affect the other. This involves using specialized parts—such as orthogonal RNA polymerases or ribosomes—that do not interact with the host's system, thereby minimizing unwanted cross-talk [30].
Q: My circuit works perfectly in bacteria, but fails in plant cells. How can I make it more portable? A: Portability failures often stem from host-specific factors. To improve cross-species function:
- Use Orthogonal Parts: Implement CRISPR-based integrators or recombinase systems (e.g., BLADE platform) that are known to function across diverse organisms [10].
- Characterize in Model Systems: First, test and optimize circuit modules in a standard model system (e.g., Nicotiana benthamiana for plants) before moving to your target organism [10].
- Employ Host-Aware Modeling: Use computational frameworks that simulate host-circuit interactions (resource competition, burden) specific to your target host to predict and mitigate issues beforehand [2].
Q: Are there ways to build "memory" into a synthetic circuit? A: Yes, irreversible memory circuits can be engineered using DNA recombinases. Serine integrases, such as PhiC31 and Bxb1, can permanently flip or invert DNA segments between their specific recognition sites. This creates a stable, heritable change in the DNA sequence that records an event (e.g., exposure to a signal) and locks the circuit in a new state, even after the signal is gone [10].
Q: What are the biggest challenges in moving synthetic gene circuits into the clinic? A: The primary translational challenges include [31]:
- Precise Control: Inability to perfectly control therapeutic activity in vivo, leading to off-target effects or immunotoxicity.
- Delivery: Difficulties in efficiently delivering genetic circuits to the correct target cells in the human body.
- Evolutionary Stability: Ensuring the circuit remains functional and non-toxic over the long term within a patient, avoiding evolutionary loss-of-function [2].
- Adaptation: Designing circuits that can dynamically respond to the complex and changing disease microenvironment.
The Scientist's Toolkit: Research Reagent Solutions
| Research Reagent | Function in Orthogonal Design | Example Use Case |
|---|---|---|
| Cre Recombinase | Edits DNA spacer length to tune promoter-gene distance, enabling post-delivery control of expression levels [3]. | Fine-tuning expression of a therapeutic transgene in human cell lines. |
| Orthogonal DNA/RNA Polymerases | Enzymes that recognize specific, engineered promoter sequences not found in the host genome, decoupling transcription [30]. | Creating multiple independent circuits in a single cell without interference. |
| Serine Integrases (e.g., PhiC31, Bxb1) | Catalyze irreversible, site-specific recombination of DNA, enabling the construction of permanent memory switches and logic gates [10]. | Recording a developmental event or environmental exposure in a plant root. |
| CRISPR-dCas9 System | Provides a programmable platform for sensors and integrators. sgRNAs guide a nuclease-deficient Cas9 (dCas9) to specific DNA sequences to repress (CRISPRi) or activate transcription [10]. | Building complex logic gates (e.g., NOR) that respond to multiple cellular inputs. |
| sRNAs (Small RNAs) | Mediate post-transcriptional regulation by binding to target mRNAs and silencing their expression. Can be used for efficient feedback control with low burden [2]. | Implementing a high-performance, burden-reducing feedback controller in bacteria. |
| Phase-Separating Proteins | Form biomolecular condensates via liquid-liquid phase separation, creating physical compartments that localize and stabilize synthetic genetic circuits [27]. | Protecting a fragile metabolic pathway from dilution during cell division in a bioproduction strain. |
Frequently Asked Questions (FAQs)
Q1: My synthetic gene circuit fails to produce any output. What are the most common causes?
A: A lack of output expression can stem from several common issues in your experimental setup [32]:
- Insufficient or Poor-Quality DNA: Using too little or degraded plasmid DNA can prevent successful transformation. Ensure you use up to 50 ng of high-quality, purified plasmid per 50 μL reaction.
- Suboptimal PCR Conditions: The wrong annealing temperature or extension time during amplification can lead to no product. Optimize annealing temperature to 5-10°C below your primer's lowest melting temperature and allow 30 seconds per 1 kb for extension.
- Poor Primer Design: Primers with strong secondary structures can hinder amplification. Use dedicated primer design tools to reduce potential secondary structures or increase primer length.
- Incorrect DNA Polymerase: Using a non-recommended polymerase can cause failure. We recommend using 1 unit of AccuPrime Pfx DNA Polymerase for amplification in mutagenesis kits.
Q2: I observe inconsistent expression of my circuit across a cell population. How can I improve uniformity?
A: Inconsistent expression often results from growth-mediated dilution, where key circuit components become diluted as cells grow and divide, leading to a loss of function [8]. A novel strategy to combat this leverages a natural cellular process called liquid-liquid phase separation. You can stabilize your circuit by fusing transcription factors (TFs) to intrinsically disordered regions (IDRs). This drives the formation of transcriptional condensates—droplet-like compartments that concentrate TFs at their target promoters, acting as molecular safe zones that buffer against dilution and preserve circuit function across cell generations [8].
Q3: What delivery method is recommended for sensitive cell types or large cargo like Cas9 ribonucleoproteins?
A: For sensitive cells or large cargo, traditional delivery methods can be harsh. Progressive Mechanoporation (PM) is a gentle, high-throughput microfluidic technique that efficiently delivers impermeable cargo [33]. It works via multistage cell deformation, combining hydrodynamic forces with rapid contact-based compression to create transient pores in the cell membrane. PM has been shown to deliver 4 kDa molecules with >90% efficiency and can even deliver functional >190 kDa Cas9 protein–sgRNA complexes without significantly affecting cell viability or proliferation rates [33].
Q4: How can I make my synthetic gene circuit less disruptive to the host cell's normal functions?
A: The key principle is orthogonality [4]. This means using genetic parts that interact strongly with each other but minimally with the host's native cellular components. To achieve this:
- Use genetic components from other organisms, such as bacterial transcription factors, bacteriophage recombinases, or CRISPR/Cas systems [4].
- These orthogonal parts reduce cross-talk with endogenous plant gene regulatory processes, minimizing metabolic burden and unintended pleiotropic effects, which is vital for predictable circuit performance [4].
Troubleshooting Guides
Table 1: Troubleshooting Gene Circuit Delivery
| Symptom | Possible Cause | Recommended Solution |
|---|---|---|
| Low delivery efficiency in compliant cells | Small pore size and short lifetime during mechanoporation [33]. | Use Progressive Mechanoporation (PM) and optimize for small molecules [33]. |
| Low delivery efficiency in stiff cells | Insufficient membrane disruption for larger cargo [33]. | Use Progressive Mechanoporation (PM) and optimize for larger molecules [33]. |
| Poor cell viability post-delivery | Excessive shear stress or compression [33]. | Implement a gentler, multistage deformation method like PM; reduce applied pressure/flow rate [33]. |
| Inability to deliver large cargo (e.g., Cas9-RNP) | Cargo size exceeds capacity of delivery method [33]. | Adopt Progressive Mechanoporation (PM), which is proven for cargo >190 kDa [33]. |
Table 2: Troubleshooting Gene Circuit Stability and Expression
| Symptom | Possible Cause | Recommended Solution |
|---|---|---|
| Circuit function lost over cell generations | Growth-mediated dilution of circuit components [8]. | Stabilize circuits using transcriptional condensates by fusing TFs to IDRs [8]. |
| Unintended effects on host cell growth | Metabolic burden or cross-talk with native processes [4]. | Re-design circuit for orthogonality using bacterial TFs, recombinases, or CRISPR/Cas parts [4]. |
| No product from PCR amplification | Poor primer design or incorrect annealing [32]. | Re-design primers using dedicated tools; optimize annealing temperature and extension time [32]. |
| DNA degradation in synthesis reaction | Overdigestion by enzymes like CorrectASE [32]. | Ensure reaction does not exceed 60 minutes and is kept on ice until the PCR step [32]. |
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for Gene Circuit Experiments
| Item | Function | Example & Notes |
|---|---|---|
| AccuPrime Pfx DNA Polymerase | High-fidelity amplification for PCR-based steps like mutagenesis [32]. | Recommended for use with GeneArt Site-Directed Mutagenesis Kits [32]. |
| Intrinsically Disordered Regions (IDRs) | Fusion partners for transcription factors to promote phase separation [8]. | Used to form transcriptional condensates that buffer against growth-mediated dilution [8]. |
| Orthogonal Genetic Parts | Reduce interference with host cell machinery [4]. | Bacterial TFs (e.g., from the TetR family), bacteriophage recombinases (e.g., Flp, B3), CRISPR/dCas9 systems [4]. |
| Dam Methylation-Sensitive Restriction Enzymes | Enzymes whose activity is blocked by dam methylation [32]. | e.g., XbaI. Use strains lacking dam methylation for cloning if digestion is inhibited [32]. |
Experimental Protocols
Detailed Protocol 1: Stabilizing a Circuit Using Transcriptional Condensates
This protocol outlines a strategy to enhance the long-term stability of synthetic gene circuits by forming protective transcriptional condensates [8].
- Design Fusion Constructs: Genetically fuse your circuit's transcription factor (TF) to an intrinsically disordered region (IDR). The choice of IDR can be optimized for different properties.
- Clone and Verify: Clone the TF-IDR fusion gene into your desired expression vector alongside the target promoter and output gene.
- Transfect/Deliver: Introduce the constructed plasmid into your target cells using an appropriate delivery method (e.g., Progressive Mechanoporation for sensitive cells).
- Validate Condensate Formation: After 24-48 hours, use microscopy to visualize the formation of bright, droplet-like condensates within the cell nucleus.
- Assess Circuit Stability: Measure the output of your circuit (e.g., fluorescence, metabolite production) over multiple cell generations and compare it to a control circuit without the IDR fusion. The stabilized circuit should maintain consistent output despite cell growth and division.
Detailed Protocol 2: Intracellular Delivery via Progressive Mechanoporation (PM)
This protocol describes using a microfluidic device for high-efficiency, gentle intracellular delivery [33].
- Device Setup: Prime a polydimethylsiloxane (PDMS)-based PM microfluidic device with an appropriate buffer.
- Prepare Cell Suspension: Harvest and resuspend your cells in a buffer containing the cargo to be delivered (e.g., Cas9-RNP, plasmids, dyes). A sample volume of about 20 μL is sufficient.
- Configure Controller: Set the pressure-based microfluidic controller to the desired operating pressure and flow rate. The device can handle high throughput (>10,000 cells per second).
- Process Cells: Flow the cell-cargo suspension through the device. Cells will experience a multistage deformation (pre-deformation followed by compression), leading to transient membrane poration and cargo uptake.
- Collect and Culture: Collect the processed cells and transfer them to a culture medium. The cells should maintain high viability and proliferation capacity.
Supporting Diagrams
Diagram 1: Synthetic Gene Circuit Module Logic
Diagram 2: Circuit Stabilization by Transcriptional Condensates
Assessing Circuit Performance, Longevity, and Real-World Applicability
Frequently Asked Questions
What do τ50 and τ±10 measure, and why are they both necessary? τ±10 (time for output to fall outside P₀ ±10%) measures short-term stability, indicating how long a circuit performs near its designed specification. τ50 (time for output to fall below P₀/2) measures long-term persistence, showing how long the circuit retains basic function. Both are crucial because a circuit might degrade quickly from its optimal state (low τ±10) but still be useful for a long time if it retains some function (high τ50) [2].
My gene circuit's output is degrading rapidly. What are the first things I should check? First, verify that your culture conditions and passaging protocol are consistent, as fluctuations can mimic evolutionary instability. Next, consider implementing a genetic controller. Research indicates that post-transcriptional controllers using small RNAs (sRNAs) generally outperform transcriptional ones, and growth-based feedback can significantly extend the circuit's functional half-life [2].
How can I experimentally measure τ50 and τ±10 for my circuit? You will need to run a long-term evolution experiment with serial passaging, tracking the total population-level output of your circuit's protein (e.g., GFP) over time [2]. The table below outlines the core measurements and calculations.
| Metric | Measurement | Typical Experimental Setup |
|---|---|---|
| Initial Output (P₀) | Total protein output from the ancestral population before mutation [2] | Measured via fluorescence or other assay at time zero. |
| Stable Output Duration (τ±10) | Time for population output P to fall outside the range P₀ ± 10% [2] | Serial passaging with daily output measurement. |
| Functional Half-Life (τ50) | Time for population output P to fall below P₀/2 [2] | Serial passaging until output drops below half of P₀. |
Troubleshooting Guides
Problem: Consistently Low τ50 (Rapid Loss of All Function) This indicates that non-functional mutants are taking over your population very quickly, often due to a high fitness cost (burden) of your circuit.
- Potential Cause 1: Excessive metabolic burden from high circuit expression.
- Solution: Implement a negative feedback controller to reduce unnecessary resource consumption. Studies show negative autoregulation can prolong short-term performance [2].
- Potential Cause 2: No evolutionary pressure to maintain the circuit.
- Solution: Couple circuit function to cell survival, for example, by linking the expression of your gene to an antibiotic resistance gene [2].
Problem: Consistently Low τ±10 (Rapid Drift from Set Point) Your circuit loses its fine-tuned function quickly, even if some output remains. This points to an accumulation of mutations that subtly alter expression levels.
- Potential Cause: Circuit is sensitive to small, function-degrading mutations.
- Solution: Use a more robust control architecture. "Host-aware" computational models suggest that multi-input controllers can improve both short- and long-term performance [2]. Furthermore, a new system called DIAL allows for post-delivery fine-tuning of gene expression levels by editing the DNA spacer between a promoter and the gene, which can help maintain a consistent set point across a population [3].
Experimental Protocols
Protocol 1: Serial Passaging Experiment for Longevity Metrics
This protocol is adapted from methodologies used to evaluate evolutionary longevity in engineered bacteria [2].
- Strain Preparation: Transform your synthetic gene circuit into the host organism (e.g., E. coli).
- Initial Measurement (P₀): Inoculate a single colony into culture medium and grow to the mid-exponential phase. Measure the total population-level output (e.g., fluorescence for a reporter protein). This value is P₀ [2].
- Serial Passaging:
- Each day, dilute the culture into fresh medium (e.g., 1:100 or 1:1000 dilution) to maintain repeated batch conditions.
- At each passage, sample and measure the total output P.
- Continue passaging for a predetermined number of generations (e.g., 100-200 generations).
- Data Analysis:
- Plot P over time (or generations).
- Calculate τ±10: Identify the time point where P first moves outside the P₀ ±10% range.
- Calculate τ50: Identify the time point where P drops below P₀/2.
Protocol 2: Implementing a DIAL Control System for Set-Point Stability
This protocol is based on a system designed to establish and edit consistent gene expression levels in a population of cells [3].
- Circuit Design:
- Design your gene circuit with a promoter, a long, excisable DNA spacer, and your gene of interest.
- Within the spacer, incorporate multiple excision sites for recombinase enzymes (e.g., lox sites for Cre recombinase).
- Delivery: Transfer the constructed DNA segment into your target cells (e.g., via viral vector).
- Set-Point Tuning:
- To increase expression from a low baseline, add the corresponding recombinase (e.g., Cre) to the cells.
- The enzyme will excise parts of the spacer, shortening the distance between the promoter and the gene, thereby increasing gene expression.
- This allows you to define "high," "med," "low," and "off" set points for your gene [3].
- Validation: Measure protein output across the cell population to confirm uniform expression at the desired level.
The Scientist's Toolkit
| Research Reagent | Function |
|---|---|
| Cre Recombinase | Enzyme used in the DIAL system to excise DNA spacer sequences, enabling post-delivery tuning of gene expression levels [3]. |
| Small RNAs (sRNAs) | Key component of post-transcriptional genetic controllers; they silence circuit mRNA, offering strong control with reduced burden [2]. |
| Fluorescent Reporter Proteins (e.g., GFP) | A standard tool for quantifying gene circuit output and tracking expression levels over time in evolution experiments [2]. |
| "Host-Aware" Computational Model | A multi-scale modeling framework that simulates host-circuit interactions, mutation, and population dynamics to predict evolutionary longevity in silico [2]. |
Workflow and Relationship Diagrams
Gene Circuit Stability Validation Workflow
Genetic Controller Architectures
Troubleshooting Guides and FAQs
FAQ 1: My synthetic gene circuit loses function after several cell generations. What are the primary causes and solutions?
Circuit failure is often caused by the metabolic burden of your circuit, which slows host cell growth and selects for non-functional mutants [2] [21]. Key vulnerabilities include:
- High metabolic burden: Circuit expression diverts host resources (ribosomes, amino acids, energy), reducing growth rate and creating a selective advantage for mutant cells with reduced circuit function [2] [1].
- Genetic instability: Repeated DNA sequences can promote recombination-mediated deletions [21]. Plasmid-based circuits are also susceptible to segregation loss during cell division [21].
- Mutation and selection: Mutations in promoters, ribosome binding sites, or coding sequences can reduce circuit function. These faster-growing mutants can overtake the population [2].
Mitigation Strategies:
- Implement genetic controllers: Use negative feedback or growth-based feedback controllers to automatically regulate circuit expression and reduce burden [2].
- Reduce genetic load: Genomically integrate the circuit to prevent plasmid loss and avoid repetitive DNA sequences [21].
- Use orthogonal parts: Employ genetic components (e.g., bacterial transcription factors) that do not interfere with host processes to minimize cross-talk and burden [4].
FAQ 2: How can I predict how long my gene circuit will remain functional in a growing cell population?
You can quantify evolutionary longevity using specific metrics in host-aware multi-scale models [2]. These models simulate host-circuit interactions, mutation, and population dynamics.
Key Longevity Metrics:
- Initial Output (P₀): Total circuit output before any mutation occurs.
- Stable Performance Duration (τ±₁₀): Time for population-wide output to fall outside P₀ ± 10%.
- Functional Half-Life (τ₅₀): Time for total output to fall below 50% of P₀ [2].
FAQ 3: What modeling framework can I use to simulate circuit evolution and host-circuit interactions?
A "host-aware" computational framework uses ordinary differential equations to model competing cell populations with different circuit mutation states [2]. The model captures:
- Resource coupling: How the circuit consumes host resources (ribosomes, metabolites) and impacts growth.
- Mutation: Transitions between strains with varying levels of circuit function (e.g., 100%, 67%, 33%, 0% of nominal output).
- Dynamic selection: Competition between strains based on their calculated growth rates in shared nutrient environments [2].
FAQ 4: My circuit output is highly variable between cells, leading to unreliable performance. How can I stabilize it?
Variability can arise from uneven gene delivery, differences in plasmid copy number, or natural cell-to-cell variation [3].
- Use a tunable expression system: Implement systems like DIAL, which uses a spacer sequence between the promoter and gene. Cre recombinase can edit this spacer to establish precise, uniform expression levels across a cell population [3].
- Employ transcriptional condensates: Fuse transcription factors to intrinsically disordered regions (IDRs) to form liquid-liquid phase-separated droplets. These condensates concentrate circuit components, buffering against dilution during cell growth and stabilizing gene expression [8].
Quantitative Data on Circuit Stability and Controller Performance
Table 1: Key Metrics for Quantifying Evolutionary Longevity in Gene Circuits [2]
| Metric | Description | Interpretation |
|---|---|---|
| P₀ | Initial total circuit output prior to mutation. | Measures the initial performance and yield of the circuit. |
| τ±₁₀ | Time for population output to fall outside P₀ ± 10%. | Indicates the duration of stable, near-nominal performance. |
| τ₅₀ | Time for population output to fall below P₀/2. | Measures functional half-life or "persistence" of the circuit. |
Table 2: Comparison of Strategies for Enhancing Circuit Longevity
| Strategy | Mechanism | Key Findings/Performance |
|---|---|---|
| Post-transcriptional Control (sRNA) | Uses small RNAs to silence circuit mRNA. | Generally outperforms transcriptional control; enables strong regulation with lower burden [2]. |
| Growth-Based Feedback | Controller senses and responds to host growth rate. | Significantly extends functional half-life (τ₅₀) in the long term [2]. |
| Negative Autoregulation | Circuit output represses its own expression. | Prolongs short-term performance (τ±₁₀) [2]. |
| Transcriptional Condensates | Phase-separated droplets concentrate circuit components. | Buffers against growth dilution, stabilizing memory in self-activation circuits [8]. |
| Multi-Input Controllers | Combines multiple feedback inputs (e.g., output and growth). | Improves circuit half-life over threefold without coupling to essential genes [2]. |
Experimental Protocols for Key Experiments
Protocol: Simulating an Evolving Population of Engineered Cells [2]
1. Objective: To model the evolutionary dynamics of a synthetic gene circuit within a bacterial population, predicting the loss of function over time.
2. Model Setup:
- Host-Circuit Interaction Model: Use a set of ordinary differential equations (ODEs) that explicitly model the consumption of host resources (e.g., ribosomes
R, metabolitese) by the circuit's mRNA (mA) and protein (pA) production [2]. - Population Model: Define multiple competing cell strains. Each strain
irepresents a different mutant with a specific maximal transcription rate (ωA_i) for the circuit gene (e.g., 100%, 67%, 33%, 0% of nominal). - Mutation Scheme: Define transition rates between strains. Only allow function-reducing mutations, with more severe mutations (e.g., from 100% to 0%) being less probable than milder ones [2].
3. Simulation Parameters:
- Growth Conditions: Simulate repeated batch culture. Replenish nutrients and reset population size every 24 hours.
- Initial State: Start the simulation with a population composed entirely of the ancestral, fully functional strain (100%
ωA). - Output Calculation: At each time point, calculate the total population output
Pas the sum ofpAacross all strains:P = Σ (N_i * pA_i), whereN_iis the number of cells in straini[2].
4. Data Analysis:
- Track the population composition over time.
- Calculate the longevity metrics (P₀, τ±₁₀, τ₅₀) from the output trajectory
P(t).
Visualization of Key Concepts and Workflows
Multi-Scale Host-Aware Modeling Framework
Mechanism of Transcriptional Condensates for Stabilization
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials and Reagents for Circuit Stability Research
| Item | Function/Description | Example Use Case |
|---|---|---|
| Host-Aware Model (in silico) | A multi-scale ODE framework simulating resource coupling, mutation, and population dynamics [2]. | Predicting evolutionary longevity (τ₅₀) of a circuit design before construction. |
| Orthogonal Genetic Parts | Components (e.g., bacterial TFs, CRISPR/Cas) that function independently of host machinery to minimize cross-talk [4]. | Building circuits that impose lower metabolic burden and are more predictable. |
| Small RNAs (sRNAs) | Non-coding RNAs used for post-transcriptional regulation of circuit mRNA [2]. | Implementing efficient negative feedback controllers with low burden. |
| Intrinsically Disordered Regions (IDRs) | Protein domains that drive liquid-liquid phase separation [8]. | Engineering transcriptional condensates to stabilize circuit components against dilution. |
| Site-Specific Recombinases (e.g., Cre) | Enzymes that catalyze precise DNA excision or inversion at specific target sites [3]. | Fine-tuning circuit expression levels by editing spacer sequences in the DIAL system. |
| Reduced-Genome Host Strains | Engineered microbes with deleted transposable elements and mobile DNA to lower background mutation rates [21]. | Hosting complex circuits to suppress the emergence of loss-of-function mutants. |
Synthetic gene circuits are engineered systems that program cells to perform desired functions, with applications ranging from therapeutic production to advanced biosensing. A fundamental challenge in their design is ensuring robust and stable expression of the circuit's genes. Two primary levels of control are utilized to achieve this: transcriptional regulation and post-transcriptional regulation. Transcriptional regulation involves controlling the initial step of gene expression, where a gene's DNA sequence is copied into RNA. This is typically achieved through transcription factors (TFs) that bind to specific promoter regions to activate or repress transcription [34]. In contrast, post-transcriptional regulation occurs after the RNA has been made, influencing its stability, localization, and translation into protein. This often involves mechanisms like microRNAs (miRNAs) that bind to messenger RNA (mRNA) to repress translation or trigger degradation [34].
The choice between these strategies significantly impacts key performance metrics of a gene circuit, including its evolutionary longevity, resistance to mutational degradation, precision of expression control, and the metabolic burden it imposes on the host cell [2]. This technical support center provides a comparative analysis and troubleshooting guide to help researchers select and optimize the right control strategy for their specific experimental needs, directly supporting thesis research on troubleshooting synthetic gene circuit expression and stability.
Quick-Reference Comparison Tables
Table 1: Performance Comparison of Regulation Strategies
| Control Strategy | Typical Actuator | Evolutionary Half-Life (Relative) | Short-Term Stability (τ±10) | Resource Burden | Ideal Use Case |
|---|---|---|---|---|---|
| Transcriptional Control | Transcription Factors (TFs) [2] | Base (1x) | Moderate | High [2] | Simple, high-output expression systems |
| Post-Transcriptional Control | Small RNAs (sRNAs), miRNAs [2] [35] | >3x improvement [2] | High | Lower [2] | Long-lasting, stable circuits; precise dosage control |
| Hybrid/Multi-Input Controllers | TFs & sRNAs combined [2] | Highest (Optimized) | Very High | Variable | Mission-critical applications requiring maximum robustness |
Table 2: Troubleshooting Common Experimental Issues
| Problem | Transcriptional Control Solution | Post-Transcriptional Control Solution |
|---|---|---|
| Rapid loss of circuit function | Implement negative autoregulation [2]. | Use sRNA-based controllers for longer half-life [2]. |
| High metabolic burden | Use weaker promoters; employ feedback loops [2]. | sRNA controllers offer strong control with less burden [2]. |
| Unwanted expression noise | N/A | The ComMAND circuit uses microRNAs for noise attenuation [35]. |
| Protein level drift during cell growth | N/A | Use phase separation (IDR fusions) to buffer against dilution [8] [12]. |
| Difficulty tuning expression level | Use a system like DIAL to edit promoter spacing post-delivery [3]. | N/A |
Experimental Protocols & Methodologies
Protocol: Distinguishing Transcriptional and Post-Transcriptional Dysregulation
Purpose: To determine whether observed differential gene expression in a disease or experimental condition is caused by changes at the transcriptional or post-transcriptional level. This is vital for identifying potential causal disease genes [36].
Background: Transcriptional regulation primarily affects the synthesis of pre-mRNA, while post-transcriptional regulation affects the processing and stability of mature mRNA. Intronic reads serve as a proxy for pre-mRNA (transcriptional activity), while exonic reads represent mature mRNA (subject to both regulation types) [36].
Methodology (Linear Mixed Model):
- RNA Sequencing & Data Preparation: Perform total RNA-seq on case and control samples. Map reads to the genome and separate reads mapping to intronic regions from those mapping to exonic regions for each gene.
- Model Fitting: For each gene, fit a linear mixed model to the expression data. The model for an expression measurement ( y{ijgk} ) is:
( y{ijgk} = Gg^T + Gg^{PT} + VG{jg}^T + VG{jg}^{PT} + Ai + \epsilon{ijgk} )
Where:
- ( Gg^T ) and ( Gg^{PT} ) are the overall mean expression levels for gene ( g ) at transcriptional and post-transcriptional levels.
- ( VG{jg}^T ) and ( VG{jg}^{PT} ) are the interaction terms of primary interest, capturing the variation in gene ( g )'s expression across treatment group ( j ) at the two regulatory levels.
- ( Ai ) is the random subject effect.
- ( \epsilon{ijgk} ) is the random error.
- Probe Region Specification:
- For a probe/read in an intron region, the model simplifies to ( y{ijgk}' = Gg^T + VG{jg}^T + Ai + \epsilon_{ijgk} ), excluding post-transcriptional effects.
- For a probe/read in an exon region, the measurement includes effects from both levels.
- Statistical Testing: Use restricted maximum likelihood (REML) estimation via the
lmer()function in the R packagelme4to fit the model. Test the null hypotheses that the effects ( VG{jg}^T ) and ( VG{jg}^{PT} ) are equal to zero using mixed-model-based t-tests. - Interpretation:
- A significant ( VG{jg}^T ) indicates differential expression at the transcriptional level.
- A significant ( VG{jg}^{PT} ) indicates differential expression at the post-transcriptional level.
- Genes with significant post-transcriptional changes are considered potential candidate causal genes for the disease, as they are more likely to alter protein expression [36].
Protocol: Implementing the DIAL System for Set-Point Control
Purpose: To achieve precise, stable, and adjustable ("dial-able") expression levels of a synthetic gene after it has been delivered into cells, overcoming variations in copy number and cellular context [3].
Background: The DIAL (set point DIALing) system controls gene expression by varying the distance between a gene and its promoter. A longer DNA "spacer" between the promoter and the gene results in lower expression, as it makes it harder for transcription factors to initiate transcription. This spacer contains sites that can be excised by recombinase enzymes, allowing for post-delivery adjustment of the spacer length and thus the expression level [3].
Methodology:
- Circuit Construction: Assemble the synthetic gene circuit where the gene of interest is separated from its promoter by a designed DNA spacer sequence.
- Incorporation of Excision Sites: Engineer the spacer to contain multiple recognition sites for site-specific recombinases (e.g., Cre recombinase). Each site represents a potential unit of length that can be removed.
- Delivery: Transfer the complete DNA construct (promoter + spacer + gene) into the target cells using a standard delivery vehicle (e.g., lentivirus, adeno-associated virus).
- Set-Point Tuning: At any time after delivery, add the corresponding recombinase enzymes to the cells. As these enzymes excise segments of the spacer, the effective distance between the promoter and gene is shortened, thereby increasing gene expression in a step-wise manner.
- Validation: Measure the output (e.g., fluorescent protein intensity) to confirm uniform expression across the cell population has been achieved at the desired "low," "med," or "high" set point [3].
Core Signaling Pathways & Workflows
Diagram: Transcriptional vs. Post-Transcriptional Controller Architecture
Diagram: Incoherent Feedforward Loop (IFFL) for Noise Control
The Scientist's Toolkit: Research Reagent Solutions
| Reagent / Resource | Type/Function | Example & Application |
|---|---|---|
| Bacterial Transcription Factors | Transcriptional Regulator | Used in orthogonal circuits to minimize host cross-talk (e.g., in plant circuits) [4]. |
| Site-Specific Recombinases | DNA Editing Enzyme | Cre recombinase; used in the DIAL system for post-delivery tuning of expression levels [3]. |
| CRISPR/Cas Components | Transcriptional Regulator / DNA Editor | Used as actuators in circuits to repress or activate endogenous genes [4]. |
| Intrinsically Disordered Regions (IDRs) | Protein Tag | Fused to TFs to drive phase separation, forming transcriptional condensates that buffer against growth-mediated dilution [8] [12]. |
| Small RNAs (sRNAs) | Post-Transcriptional Regulator | Key actuator in post-transcriptional controllers; provides strong, low-burden regulation [2]. |
| microRNAs (miRNAs) | Post-Transcriptional Regulator | Core component of the ComMAND IFFL circuit; represses translation to attenuate noise and control dosage [35]. |
| Inducible Promoters | Sensor Module | Respond to inputs like dexamethasone, β-Estradiol, or copper ions; used to drive circuit components in response to stimuli [4]. |
| TRANSFAC / miRTarBase | Database | Curated databases of transcription factor binding sites and experimentally validated miRNA-target interactions [34]. |
Troubleshooting Guides and FAQs
Frequently Asked Questions
Q1: My synthetic gene circuit loses function after a few cell divisions. What could be causing this?
A: This is typically caused by evolutionary instability [2]. Your circuit imposes a metabolic burden on host cells, slowing their growth. Mutant cells with non-functional, less burdensome circuits will outcompete the original engineered cells [2]. To mitigate this:
- Implement Genetic Controllers: Use feedback controllers that sense host-circuit interactions to maintain stable expression. Post-transcriptional controllers using small RNAs (sRNAs) can be particularly effective [2].
- Employ Phase Separation: Fuse transcription factors to intrinsically disordered regions (IDRs) to form transcriptional condensates. These "molecular safe zones" concentrate key circuit components, buffering them against growth-mediated dilution [8] [12].
Q2: How can I achieve uniform and consistent expression of my synthetic gene across a entire population of cells?
A: Inconsistent expression often stems from variations in copy number and natural cell-to-cell differences [3]. The DIAL (Distance-Induced Adjustment of Levels) system directly addresses this [3].
- Principle: The system uses a DNA "spacer" between the promoter and the gene. A longer spacer reduces gene expression. Enzymes like Cre recombinase can be added later to excise parts of this spacer, bringing the promoter closer and "dialing up" expression in a controlled manner [3].
- Protocol: Design your gene circuit with the DIAL spacer sequence. After transferring the circuit into cells, you can add specific recombinases to uniformly adjust the expression set point across the entire cell population to a desired "high," "med," or "low" level [3].
Q3: The output of my biosensing living material is inconsistent under real-world conditions. How can I improve its reliability?
A: Traditional whole-cell biosensors are sensitive to environmental fluctuations [37]. Integration into Engineered Living Materials (ELMs) enhances stability.
- Solution: Embed your engineered sensing cells within a synthetic matrix, such as a hydrogel [37]. The matrix protects the cells from environmental interference and contaminants, while also providing mechanical stability. Furthermore, you can design genetic circuits within the cells to respond to a wider range of stimuli (e.g., light, mechanical stress) for more robust sensing applications [37].
Troubleshooting Guide: Common Problems and Solutions
| Problem | Possible Cause | Recommended Solution |
|---|---|---|
| Rapid loss of circuit function | Evolutionary selection for low-burden mutants [2] | Implement growth-based negative feedback controllers or couple circuit function to an essential gene [2]. |
| High cell-to-cell variability in output | Variation in circuit copy number; biological noise [3] | Use the DIAL system to post-transcriptionally fine-tune and unify expression levels across the population [3]. |
| Unstable memory in toggle switches or logic gates | Growth-mediated dilution of transcription factors [12] | Redesign circuits using phase separation; fuse TFs with IDRs to form stabilizing condensates [8] [12]. |
| Poor performance in environmental biosensing | Fluctuating temperature, pH, or biological contaminants [37] | Integrate sensing cells into a protective hydrogel-based ELM to enhance resilience [37]. |
| Low yield in bioproduction circuits | Metabolic burden; resource competition with host [2] | Apply "host-aware" design principles and use controllers that optimize resource allocation without completely shutting down production [2]. |
Experimental Data and Protocols
Performance Metrics of Selected Synthetic Gene Circuits
The table below summarizes the performance of various synthetic gene circuits, highlighting their inputs, outputs, and key operational metrics, which are crucial for selecting the right design for your application.
Table 1: Circuit Performance in Diverse Applications [37]
| Stimulus Type | Input Signal | Output Signal | Host Organism | Material | Threshold | Functional Stability |
|---|---|---|---|---|---|---|
| Synthetic Inducer | IPTG | RFP (fluorescence) | E. coli | Hydrogel | 0.1–1 mM | >72 hours |
| Environmental Chemical | Pb²⁺ | mtagBFP (fluorescence) | B. subtilis | Biofilm@biochar | 0.1 μg/L | >7 days |
| Light | Blue Light (470 nm) | NanoLuc (luminescence) | S. cerevisiae | Bacterial Cellulose | N/A | >7 days |
| Heat | >39 °C | mCherry (fluorescence) | E. coli | GNC Hydrogel | 39 °C | Not explicitly quantified |
| Mechanical Loading | 15% compressive strain | IL-1Ra (therapeutic protein) | Chondrocytes | Agarose Hydrogel | 15% strain | ≥3 days |
Detailed Experimental Protocol: Stabilizing Circuits with Phase Separation
This protocol is adapted from research that used phase separation to protect a self-activation (SA) circuit from growth-mediated dilution [8] [12].
Objective: To enhance the resilience of a synthetic gene circuit by forming transcriptional condensates around its transcription factor.
Materials:
- Plasmids: Circuit plasmid containing the self-activation gene (e.g., a transcription factor like LuxR or TetR under its own promoter).
- Cloning Reagents: Molecular biology kits for PCR, digestion, and ligation.
- IDR Sequence: DNA sequence coding for an intrinsically disordered region (e.g., from FUS or other phase-separating proteins).
- Host Cells: E. coli or other target chassis.
- Inducers: Specific chemical inducers for your circuit (e.g., AHL for LuxR).
- Microscopy Equipment: For visualizing condensate formation (e.g., confocal microscope).
Methodology:
- Circuit Redesign: Fuse the DNA sequence of the chosen IDR to the gene encoding the transcription factor in your SA circuit. Ensure the fusion protein is correctly expressed and functional.
- Transformation: Introduce the redesigned plasmid into your host cells.
- Induction and Culture: Grow transformed cells and induce circuit activity with the appropriate chemical signal. Continue culturing through multiple growth cycles to simulate prolonged dilution.
- Validation and Analysis:
- Microscopy: Image the cells to confirm the formation of bright, droplet-like transcriptional condensates at the promoter sites.
- Output Measurement: Quantify the circuit's output (e.g., fluorescence from a reporter gene) over time and compare it to a control circuit without the IDR fusion.
- Stability Assessment: Measure how long the bistable memory or output signal is maintained in both the experimental and control groups during continuous culture.
The Scientist's Toolkit: Key Research Reagents
Table 2: Essential Reagents for Circuit Stability Research
| Reagent / Tool | Function | Application Example |
|---|---|---|
| Cre Recombinase (in DIAL system) | Edits DNA spacer sequences to fine-tune the distance between a promoter and its gene [3]. | Precisely "dial" gene expression to a desired set point after circuit delivery for uniform output [3]. |
| Intrinsically Disordered Regions (IDRs) | Mediate liquid-liquid phase separation to form biomolecular condensates [8] [12]. | Fuse to transcription factors to create localized "hubs" that resist growth-mediated dilution, stabilizing circuit memory [12]. |
| Small RNAs (sRNAs) | Enable post-transcriptional regulation by silencing target mRNAs [2]. | Implement efficient negative feedback controllers with lower burden than transcriptional controllers to enhance evolutionary longevity [2]. |
| Hydrogel Matrices (e.g., Pluronic F127-BUM, Agarose) | Synthetic, porous scaffolds that provide a 3D support structure for living cells [37]. | Create Engineered Living Materials (ELMs) by encapsulating sensor cells, protecting them from environmental stresses and improving biosensor robustness [37]. |
| "Host-Aware" Computational Model | A multi-scale framework simulating host-circuit interactions, mutation, and mutant competition [2]. | Predict the evolutionary half-life of a circuit design in silico and identify optimal controller architectures before costly wet-lab experiments [2]. |
Signaling Pathways and Workflows
Diagram: Strategies for Stabilizing Synthetic Gene Circuits
This diagram illustrates the core mechanisms behind three advanced stabilization techniques discussed in the guides.
Diagram: Engineered Living Material (ELM) Biosensing Workflow
This workflow shows how synthetic gene circuits are integrated into materials to create robust biosensors.
Conclusion
Ensuring the stable expression and evolutionary longevity of synthetic gene circuits is paramount for their successful translation into reliable biomedical tools. The integration of tunable control systems, such as the DIAL platform for set-point establishment, with physical stabilization methods, like transcriptional condensates, represents a powerful synergy of genetic and physical design principles. Furthermore, adopting a 'host-aware' perspective that accounts for metabolic burden and evolutionary pressures from the outset is crucial. Future efforts must focus on standardizing parts and characterization data, developing more sophisticated multi-scale models, and creating generalized design rules that are robust across different cellular contexts. By systematically addressing these challenges, the field can advance from demonstrating circuit function in model systems to deploying robust, predictable, and durable genetic programs for next-generation therapeutics and diagnostics.
References
- 1. How to entertain a gene circuit without exhausting its host [https://fullcircle.asu.edu/faculty/how-to-entertain-a-gene-circuit-without-exhausting-its-host/]
- 2. Genetic controllers for enhancing the evolutionary ... [https://www.nature.com/articles/s41467-025-63627-4]
- 3. A new system can dial expression of synthetic genes up or ... [https://news.mit.edu/2025/new-system-can-dial-expression-synthetic-genes-up-down-1013]
- 4. The switch‐liker's guide to plant synthetic gene circuits - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC11887007/]
- 5. Designing and engineering evolutionary robust genetic circuits [https://jbioleng.biomedcentral.com/articles/10.1186/1754-1611-4-12]
- 6. How to entertain a gene circuit without exhausting its host [https://news.asu.edu/20220225-how-entertain-gene-circuit-without-exhausting-its-host]
- 7. Dynamical mechanisms of growth-feedback effects on ... [https://elifesciences.org/articles/89170]
- 8. Stabilizing Synthetic Gene Circuits Using Transcriptional ... [https://www.genengnews.com/topics/bioprocessing/stabilizing-synthetic-gene-circuits-using-transcriptional-condensates/]
- 9. Evolutionary regain of lost gene circuit function - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6911209/]
- 10. Synthetic gene circuits in plants: recent advances and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11894408/]
- 11. Synthetic gene circuit evolution: Insights and opportunities at ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11330362/]
- 12. Phase separation to buffer growth-mediated dilution in ... [https://www.sciencedirect.com/science/article/abs/pii/S0092867425011821]
- 13. Topology-Dependent Interference of Synthetic Gene Circuit ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7246135/]
- 14. Effects of growth feedback on gene circuits: A dynamical ... [https://elifesciences.org/reviewed-preprints/89170v1]
- 15. Phase Separation to Resolve Growth-Related Circuit Failures [https://pmc.ncbi.nlm.nih.gov/articles/PMC11565989/]
- 16. Programmable promoter editing for precise control of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11212971/]
- 17. Negative autoregulation linearizes the dose–response and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2654390/]
- 18. Applied Biosystems SeqStudio Genetic Analyzer Support— ... [https://www.thermofisher.com/us/en/home/technical-resources/technical-reference-library/capillary-electrophoresis-instruments-support-center/seqstudio-genetic-analyzer-support/seqstudio-genetic-analyzer-support-troubleshooting.html]
- 19. 2. Design principles governing the rate of gene expression [https://biocircuits.github.io/chapters/02_response_time.html]
- 20. Balancing a genetic toggle switch by real-time feedback ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5693866/]
- 21. Design patterns for engineering genetic stability - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC8294169/]
- 22. Addressing biological uncertainties in engineering gene ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4837052/]
- 23. Logic Synthesis of Recombinase-Based Genetic Circuits [https://www.nature.com/articles/s41598-017-07386-3]
- 24. Scaling Computation and Memory in Living Cells - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6003718/]
- 25. Ten future challenges for synthetic biology - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9996719/]
- 26. Transcriptional condensates: a blessing or a curse for gene ... [https://www.nature.com/articles/s42003-024-05892-5]
- 27. Using nature's playbook to keep engineered cells on script [https://news.asu.edu/20251110-science-and-technology-using-natures-playbook-keep-engineered-cells-script]
- 28. conservation of regulated Evolutionary assurance... longevity [https://genomebiology.biomedcentral.com/articles/10.1186/gb-2007-8-7-r132]
- 29. Synthetic gene circuits that selectively target RAS-driven ... [https://elifesciences.org/reviewed-preprints/104320]
- 30. #063 - Orthogonal Design Principles in Engineering [https://flocode.substack.com/p/orthogonal-design-principles-in-engineering]
- 31. Translating synthetic gene circuits into the clinic [https://www.sciencedirect.com/science/article/abs/pii/S2405471225002583]
- 32. Gene Synthesis and Mutagenesis Support - Troubleshooting [https://www.thermofisher.com/us/en/home/technical-resources/technical-reference-library/cloning-technical-support-center/gene-syn-mut-cloning/gene-synthesis-and-mutagenesis-cloning-troubleshooting.html]
- 33. Efficient and gentle delivery of molecules into cells with ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8204113/]
- 34. Introductory Chapter: A Brief Overview of Transcriptional ... [https://www.intechopen.com/chapters/62601]
- 35. Gene circuits enable more precise control of gene therapy [https://news.mit.edu/2025/gene-circuits-enable-more-precise-control-gene-therapy-0428]
- 36. Identify gene expression pattern change at transcriptional and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6602563/]
- 37. Synthetic Gene Circuits Enable Sensing in Engineered Living ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12467368/]