Transcription Factor-Based Biosensors for Dynamic Regulation: From Design Principles to Biomedical Applications
This article provides a comprehensive overview of transcription factor (TF)-based biosensors as powerful tools for dynamic regulation in synthetic biology and metabolic engineering.
Transcription Factor-Based Biosensors for Dynamic Regulation: From Design Principles to Biomedical Applications
Abstract
This article provides a comprehensive overview of transcription factor (TF)-based biosensors as powerful tools for dynamic regulation in synthetic biology and metabolic engineering. Aimed at researchers, scientists, and drug development professionals, it explores the fundamental mechanisms of these genetically encoded devices, detailing how they convert small-molecule detection into precise control of cellular processes. The content covers established and emerging engineering strategies—from promoter and ribosome binding site tuning to advanced computational design—for optimizing biosensor performance parameters such as dynamic range, specificity, and sensitivity. Furthermore, it examines critical applications in high-throughput screening and dynamic pathway control, discusses validation frameworks and comparative analyses with other biosensor types, and concludes with future perspectives on integrating artificial intelligence and cell-free systems to overcome current limitations in bioproduction and therapeutic development.
The Core Mechanics of TF-Based Biosensors: Understanding Nature's Molecular Switches
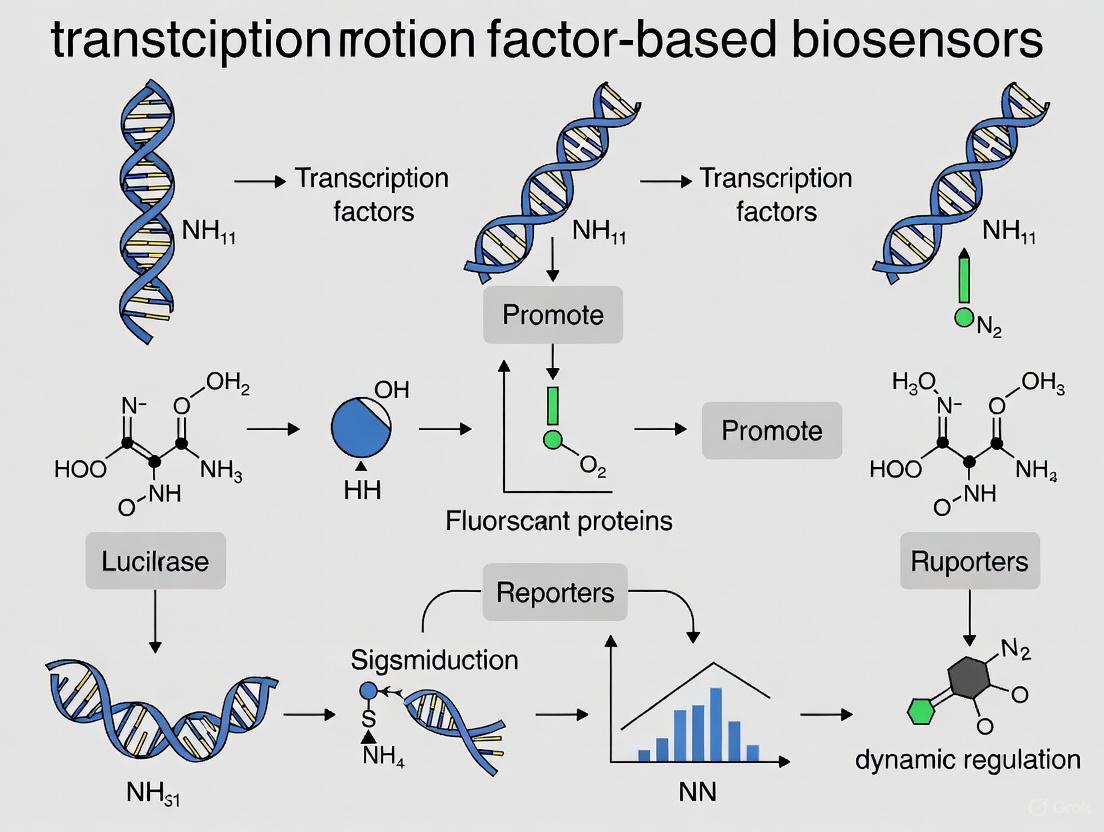
Transcription factor (TF)-based biosensors are genetically encoded tools that enable engineered cells to detect specific small molecules and convert this recognition into a measurable biological output [1] [2]. These systems are fundamental to advancing synthetic biology, serving as critical components for dynamic metabolic regulation, high-throughput screening of microbial cell factories, and real-time monitoring of metabolic fluxes [3] [4]. Their operation mirrors conventional biosensor architecture, comprising three core modules: a sensing module for analyte recognition, a transduction module for signal conversion, and an output module for generating a quantifiable signal [5]. This guide details the basic components and working principles of these modules, providing a technical foundation for their application in dynamic regulation research.
Core Component 1: The Sensing Module
The sensing module is responsible for the specific detection of an intracellular or extracellular analyte. Its core component is the transcription factor (TF), a protein that binds a target molecule (ligand) and undergoes a conformational change that modulates its ability to regulate transcription [1] [5].
Mechanism of Analyte Recognition
Transcription factors are typically modular proteins consisting of two primary domains:
- Effector-Binding Domain (EBD): This domain is responsible for ligand recognition and specificity. Upon binding its cognate ligand, the EBD undergoes a conformational change [2] [5].
- DNA-Binding Domain (DBD): This domain recognizes and binds to a specific DNA sequence, known as the operator or transcription factor binding site (TFBS), within a promoter region [2] [5].
The binding of the ligand to the EBD allosterically affects the DNA-binding affinity of the DBD. This mechanism varies depending on whether the TF functions as a repressor or an activator [5]:
- Transcriptional Repressors: In the absence of the inducer molecule, the repressor TF is bound to its operator site, physically blocking RNA polymerase (RNApol) and preventing transcription. Ligand binding induces a conformational change that causes the repressor to dissociate from the DNA, allowing transcription to proceed [5].
- Transcriptional Activators: These TFs typically bind to their operator site and recruit RNApol to the promoter only when the ligand is bound. The ligand-induced conformational change enables productive interactions with the RNApol, initiating transcription [5].
Table 1: Major Transcription Factor Families and Their Analytes
| TF Family | Primary Analyte Types | Example TFs | Regulatory Role |
|---|---|---|---|
| MerR | Metal ions, antibiotics | MerR, ArsR | Activator/Repressor |
| AraC/XylS | Aromatic compounds | XylS, AraC | Activator |
| LysR | Aromatic compounds, metabolites | LysR | Activator |
| TetR | Antibiotics, diverse small molecules | TetR | Repressor |
| MarR | Antibiotics, phenolic compounds | MarR | Repressor |
Core Component 2: The Transduction Module
The transduction module transmits the signal from the sensing module to the gene expression machinery, ultimately controlling the production of the output signal. This module is primarily composed of the TF-specific promoter [4].
The Promoter as a Signal Transducer
The promoter is a DNA sequence located upstream of the output gene. The key element for transduction is the Transcription Factor Binding Site (TFBS), the specific DNA sequence recognized by the TF's DBD [4]. The interaction between the ligand-bound TF and its TFBS determines the rate of transcription initiation for the downstream output gene. The design of this promoter, including the number, affinity, and precise location of the TFBS relative to the core promoter elements (e.g., -35 and -10 boxes), is critical for defining the performance of the biosensor [6].
Core Component 3: The Output Module
The output module is responsible for generating a quantifiable signal based on the transcriptional activity of the promoter. This allows researchers to infer the concentration of the target analyte.
Common Reporter Outputs
- Fluorescent Proteins (e.g., GFP, eGFP): Enable real-time, non-destructive monitoring of analyte concentration via fluorescence intensity measurements [1] [7].
- Luminescent Enzymes (e.g., Luciferase): Produce light as a readout, offering high sensitivity and a broad dynamic range [5].
- Enzymatic Reporters (e.g., β-Galactosidase): Catalyze reactions that generate colorimetric or chemiluminescent products, suitable for endpoint assays [5].
The output module's choice depends on the application, with fluorescent proteins being favored for high-throughput screening and real-time monitoring within live cells [1].
Performance Characterization of TF-Based Biosensors
The performance of a TF-based biosensor is evaluated using a set of key metrics, typically characterized by an input-output dose-response curve [3] [6]. The following diagram illustrates the core workflow and performance parameters.
Diagram 1: Biosensor workflow and key performance metrics
Table 2: Key Performance Metrics for Biosensor Evaluation
| Performance Metric | Definition | Impact on Biosensor Function |
|---|---|---|
| Specificity | The ability to distinguish the target analyte from other similar molecules [6]. | Reduces false positives and ensures output accuracy. |
| Sensitivity | The minimal change in analyte concentration required to produce a detectable change in output signal [6]. | Determines the biosensor's ability to detect subtle concentration changes. |
| Dynamic Range | The fold-change between the maximal and minimal output signal levels [1] [6]. | A larger dynamic range provides a clearer distinction between induced and uninduced states. |
| Operating Range | The concentration window of the analyte over which the biosensor responds effectively [3]. | Defines the useful detection limits for the biosensor. |
| Response Time | The speed at which the biosensor reaches its half-maximal output after analyte exposure [3] [6]. | Critical for applications requiring real-time monitoring and rapid feedback control. |
Essential Methodologies for Biosensor Implementation and Tuning
Constructing and optimizing a TF-based biosensor for a specific research context requires a structured experimental approach. The process typically involves genetic circuit construction, performance characterization, and iterative tuning.
Genetic Circuit Construction and Validation
The foundational step involves assembling the genetic components into a functional biosensor within a host organism (e.g., E. coli).
Detailed Protocol: Construction of a TtgR-based Flavonoid Biosensor [7]
- Component Amplification: Amplify the gene encoding the transcription factor (e.g.,
ttgRfrom Pseudomonas putida genomic DNA) and its native promoter/operator region (PttgABC) using polymerase chain reaction (PCR). - Plasmid Assembly: Clone the
ttgRgene into a plasmid with an appropriate antibiotic resistance marker (e.g., pCDF-Duet, spectinomycin resistance). Clone thePttgABCpromoter upstream of a reporter gene (e.g.,egfp) in a compatible plasmid (e.g., pET-21a(+), ampicillin resistance). - Host Transformation: Co-transform both plasmids into a suitable microbial host, such as E. coli BL21(DE3).
- Biosensor Assay:
- Inoculate overnight cultures of the biosensor strain.
- Dilute cultures into fresh medium and expose them to a range of analyte concentrations (e.g., flavonoids like naringenin or quercetin).
- Incubate with shaking to allow for gene expression.
- Measure the output signal (e.g., fluorescence intensity for eGFP) using a microplate reader or flow cytometry.
- Data Analysis: Plot the dose-response curve (output signal vs. analyte concentration) to determine initial performance metrics like dynamic range and sensitivity.
Biosensor Tuning Strategies
Native biosensors often require optimization to achieve desired performance. Tuning can be applied at multiple levels:
- Promoter Engineering: Modifying the TF-specific promoter by altering the number, sequence, or position of the TF binding sites (TFBS) to fine-tune sensitivity, dynamic range, and cooperativity [6] [2].
- Ribosome Binding Site (RBS) Engineering: Varying the strength of the RBS upstream of the TF or the reporter gene to control translation efficiency, thereby adjusting the dynamic range and output signal intensity [6] [4].
- Transcription Factor Engineering: Using directed evolution or rational design to mutate the TF's ligand-binding domain, altering its specificity, sensitivity, or operational range [6] [2] [7]. For example, engineering a TtgR variant (N110F) altered its sensing profile for flavonoids like resveratrol [7].
Research Reagent Solutions
The following table lists key reagents and materials essential for developing and working with TF-based biosensors.
Table 3: Essential Research Reagents and Materials
| Reagent/Material | Function/Application | Specific Examples |
|---|---|---|
| Transcription Factors | Core sensing element; determines analyte specificity. | TtgR (flavonoids), ArsR (arsenic), ZntR (zinc/cadmium), LuxR (acyl-homoserine lactones) [1] [7]. |
| Reporter Genes | Generates measurable output signal. | Fluorescent proteins (eGFP, Venus), luminescent enzymes (luciferase), chromogenic enzymes (LacZ) [7] [5]. |
| Expression Vectors | Plasmid backbone for hosting biosensor genetic circuits. | pCDF-Duet, pET-21a(+), pZnt-eGFP; chosen for copy number and compatibility [7]. |
| Host Strains | Microbial chassis for biosensor operation. | E. coli BL21(DE3), E. coli DH5α [7]. |
| High-Throughput Screening Equipment | Enables rapid screening of mutant libraries or analyte detection. | Flow Cytometry (for fluorescence-activated cell sorting), Microplate Readers [3] [1]. |
| Analytes/Inducers | Used for biosensor calibration and functional testing. | Flavonoids (naringenin, quercetin), heavy metals (Hg²âº, As³âº), antibiotics, metabolic intermediates [1] [7]. |
Activator-Based vs. Repressor-Based Systems
Transcription factor (TF)-based biosensors are indispensable tools in synthetic biology and metabolic engineering, enabling real-time monitoring of metabolites and dynamic control of genetic circuits. The core of these biosensors lies in their mode of action, primarily determined by whether the transcription factor functions as an activator or a repressor. Activator-based systems enhance gene expression upon sensing a target ligand, while repressor-based systems reduce it. Within the broader context of developing advanced biosensors for dynamic regulation research, understanding the distinct characteristics, advantages, and limitations of these two systems is paramount for designing efficient microbial cell factories and high-throughput screening platforms [8] [4]. This guide provides a technical comparison of these systems, supplemented with quantitative data, experimental protocols, and visual resources tailored for researchers and scientists in drug development and related fields.
Fundamental Mechanisms of Action
The operational principle of both systems hinges on allosteric transcription factors (aTFs) that undergo a conformational change upon binding a specific effector molecule (ligand). This conformational change alters the TF's ability to bind its target DNA sequence, thereby modulating transcription of the downstream gene [9] [10].
- Activator-Based Systems: In the absence of the inducer, the activator TF is typically in an inactive state and cannot bind DNA. When the ligand is present, it binds to the TF, inducing a conformational change that allows the TF to bind its cognate promoter. This binding facilitates the recruitment of RNA polymerase (RNAP), leading to the activation of gene expression [11]. A simple analogy is an activator "unlocking" a promoter to allow transcription to proceed.
- Repressor-Based Systems: Conversely, in a repressor-based system, the TF is often active in its apo state and binds to the operator site within the promoter, physically blocking RNAP from initiating transcription. Binding of the effector molecule causes the repressor to dissociate from the DNA, thereby derepressing gene expression and allowing transcription to occur [11]. This is akin to a repressor "guarding" a promoter until the key ligand tells it to step aside.
It is crucial to note that some TFs exhibit duality, meaning they can function as activators for some genes and repressors for others. Furthermore, recent research identifies incoherent TFs, which can simultaneously exert both activating and repressive effects on a single target gene, potentially leading to non-monotonic, concentration-dependent responses [12].
The following diagram illustrates the fundamental mechanisms of activator-based and repressor-based biosensor systems.
Performance Characteristics and Quantitative Comparison
The choice between an activator and a repressor system depends heavily on the required performance characteristics, which include dynamic range, sensitivity, and the basal expression level (leakiness). These parameters are typically derived from an input-output response curve fitted using the Hill equation [4].
Key Performance Metrics:
- Dynamic Range: The fold difference between the fully induced (ON) and the uninduced (OFF) states of the biosensor. A wider dynamic range is generally preferred for clear signal distinction.
- Sensitivity (EC50/IC50): The ligand concentration required to achieve half of the maximum response. For activators, this is the half-maximal effective concentration (EC50); for repressors, it is the half-maximal inhibitory concentration (IC50).
- Operating Range: The span of ligand concentrations over which the biosensor responds.
- Basal Expression (Leakiness): The level of output signal in the absence of the inducing ligand.
Table 1: Comparative Performance Characteristics of Activator vs. Repressor Systems
| Feature | Activator-Based System | Repressor-Based System |
|---|---|---|
| Default State (No Ligand) | Low output (OFF) | High output (ON) |
| State with Saturating Ligand | High output (ON) | Low output (OFF) |
| Typical Promoter Strength | Often uses weak promoters that require activation for significant transcription [11] | Often uses strong promoters that are constitutive unless repressed [11] |
| Basal Expression (Leakiness) | Can be very low | Can be high if repression is incomplete |
| Dynamic Range | Can be very high if basal expression is minimized | Can be high if repression is strong and leakiness is low |
| Common Applications | Dynamic upregulation of pathway genes; detection of metabolite accumulation [4] | High-throughput screening of producers; repression of competing pathways [9] |
Table 2: Example Biosensors and Their Modes of Action
| Transcription Factor | Effector/Ligand | Mode of Action | Host Organism | Primary Application |
|---|---|---|---|---|
| LuxR | Acyl-homoserine lactones (AHLs) | Activator | E. coli | Quorum sensing, microbial communication [8] |
| MerR / ArsR | Hg²⺠/ As³⺠| Activator | E. coli | Heavy metal detection in environmental monitoring [8] |
| FadR | Fatty acyl-CoAs | Repressor | S. cerevisiae, E. coli | Screening for strains with enhanced fatty acid pools [4] |
| TetR | Tetracycline | Repressor | E. coli | Dynamic regulation, gene circuit control [9] |
| Phenolic Acid-responsive TFs | Protocatechuic acid, others | Activator | E. coli, P. putida | High-throughput screening of enzyme variants [13] |
Experimental Protocols for Biosensor Implementation
Protocol: Construction and Validation of a Novel TFB
This protocol outlines the steps for developing and testing a new transcription factor-based biosensor, applicable to both activator and repressor systems.
Part Identification and Selection:
- Transcription Factor (TF): Identify a TF that responds to your ligand of interest. This can be done by mining databases like RegulonDB, P2TF, or PRODORIC (see The Scientist's Toolkit) [9] [10]. If a native TF is unavailable, explore homologous TFs from other species or employ directed evolution to alter the specificity of an existing TF [9] [4].
- Promoter: Identify the cognate promoter sequence containing the specific TF binding site (TFBS). The promoter's core strength (weak for activators, strong for repressors) is a key design consideration [11].
- Reporter Gene: Select a reporter gene suitable for high-throughput screening, such as GFP (fluorescence), LacZ (colorimetry), or an antibiotic resistance gene.
Genetic Circuit Assembly:
- Assemble the genetic circuit in your desired host chassis (e.g., E. coli, S. cerevisiae). The standard architecture is: TF gene (constitutively expressed) + Cognate promoter → Reporter gene.
- The TF can be expressed on the same plasmid as the reporter or on a separate plasmid. Ensure the TF gene is under a constitutive promoter with appropriate strength to avoid toxicity or insufficient sensing.
Characterization and Parameterization:
- Cultivation: Grow multiple cultures of the biosensor strain, each exposed to a different, defined concentration of the ligand.
- Measurement: Measure the output signal (e.g., fluorescence) and the cell density (OD600) for each culture during the mid-exponential growth phase.
- Data Analysis: Normalize the output signal to cell density. Plot the normalized output against the ligand concentration and fit the data to the Hill equation to determine the dynamic range, EC50/IC50, and other key parameters [13] [4].
Protocol: High-Throughput Screening of Enzyme Variants
This methodology leverages a biosensor to screen a library of enzyme variants for improved activity [13].
- Library Creation: Generate a diverse library of enzyme variants, for example, via error-prone PCR or site-saturation mutagenesis of a key pathway enzyme.
- Biosensor Coupling: Co-express or transform the enzyme variant library into the host strain containing the biosensor circuit. The biosensor must be designed to detect the product of the enzymatic reaction.
- Screening and Sorting:
- Incubate the library to allow enzyme expression and metabolite production.
- Use a fluorescence-activated cell sorter (FACS) to isolate cells exhibiting the highest fluorescence intensity, indicating high product concentration and thus high enzyme activity.
- Validation: Isolate the sorted cells, recover them, and validate the performance of the enriched enzyme variants using more precise, low-throughput methods like HPLC.
Essential Research Reagent Solutions
The following table details key reagents, databases, and tools essential for research on transcription factor-based biosensors.
Table 3: The Scientist's Toolkit for TFB Research
| Item / Reagent | Function / Description | Example Sources / Tools |
|---|---|---|
| Allosteric Transcription Factors (aTFs) | Core sensing component; binds ligand and regulates DNA binding. | Families: TetR, LysR, AraC [9]. Sources: Native hosts, metagenomic mining [10]. |
| Cognate Promoters | DNA part containing the TF binding site; controls reporter/output gene expression. | Identified via genomic studies; often sourced from the same operon as the native TF [13]. |
| Reporter Genes | Generates measurable output (e.g., fluorescence, luminescence). | GFP, RFP, LacZ, Luciferase [8] [13]. |
| TF and Promoter Databases | Curated repositories for identifying and characterizing TF-promoter pairs. | RegulonDB (E. coli), P2TF (Prokaryotes), JASPAR (TF binding profiles) [9] [10]. |
| Computational Prediction Tools | AI and bioinformatics tools for predicting new TFs, optimizing circuits, and modeling. | DeepTFactor (TF prediction), Cello (genetic circuit design), AlphaFold (protein structure prediction) [10] [8]. |
| Directed Evolution Platforms | Methods to engineer TFs with altered ligand specificity or improved performance. | Error-prone PCR, site-directed mutagenesis, and high-throughput screening [9] [4]. |
Both activator-based and repressor-based systems offer distinct advantages for constructing dynamic regulatory networks in synthetic biology. The choice between them is not merely binary but depends on the specific application, desired performance metrics, and host context. Activators are often preferred for tightly controlled, inducible expression from weak promoters, while repressors are powerful for turning off constitutive expression in response to metabolic signals. Future research will focus on expanding the library of well-characterized TFs, refining engineering strategies to minimize cross-talk and leakiness, and integrating multiple sensory modalities. The continued development of these sophisticated genetic control systems, powered by computational design and AI, is fundamental to advancing dynamic regulation research and the efficient production of biofuels, pharmaceuticals, and other high-value chemicals [14] [8] [4].
Transcription factor-based biosensors (TFBs) are indispensable tools in synthetic biology and metabolic engineering, enabling real-time monitoring of metabolites and dynamic control of metabolic pathways. Their performance in these roles is governed by three core quantitative metrics: sensitivity, dynamic range, and specificity [15]. Optimizing these parameters is critical for developing robust biosensors capable of precise, high-throughput screening and reliable regulation in complex biological systems [3] [4]. This guide details the definition, measurement, and enhancement of these key metrics for researchers and drug development professionals.
Core Performance Metrics Explained
The performance of a transcription factor-based biosensor is typically characterized by its dose-response curve, which plots the output signal (e.g., fluorescence) against the input ligand concentration [3] [4]. Key metrics are derived from this curve to form a complete performance profile.
Table 1: Definition and Significance of Core TFB Performance Metrics
| Performance Metric | Formal Definition | Interpretation on Dose-Response Curve | Impact on Biosensor Application |
|---|---|---|---|
| Sensitivity [15] | The concentration change of a metabolite required to produce a measurable change in the biosensor's output signal. | The steepness of the curve's linear phase; a steeper slope indicates higher sensitivity. | Determines the ability to detect subtle fluctuations in metabolite levels; crucial for fine dynamic regulation. |
| Dynamic Range [1] [15] | The fold-change between the maximal output signal (at saturating ligand) and the minimal basal signal (without ligand). | The difference between the upper and lower plateau of the sigmoidal curve. | Defines the resolution for distinguishing high-performing microbial strains and the amplitude of regulatory output. |
| Specificity [15] | The difference in output signal intensity upon binding the target ligand compared to alternative, non-target ligands. | Measured by comparing dose-response curves for the target ligand versus structural analogs. | Reduces false positives in screening and ensures the biosensor responds only to the intended metabolic signal. |
Other critical metrics include the operating range (the concentration window of effective operation) and the response time (the speed to reach half-maximal output) [3]. The limit of detection (LoD) is also a key parameter, representing the lowest ligand concentration that can be reliably distinguished from background noise [1].
Quantification and Experimental Protocols
Accurately quantifying these metrics requires standardized experimental workflows and data analysis.
Standardized Measurement Workflow
A general protocol for characterizing TFB performance involves the following steps [10] [16]:
- Strain Preparation: Clone the biosensor genetic circuit (TF, promoter, reporter gene) into the host microbial chassis.
- Culture and Induction: Grow cultures and expose them to a defined concentration gradient of the target ligand. A minimum of eight data points across the expected concentration range is recommended [15].
- Signal Measurement: In the log or early stationary phase, measure the output signal (e.g., fluorescence intensity, luminescence) for each culture using a plate reader.
- Data Normalization: Normalize the output signal (e.g., fluorescence) to cell density (e.g., OD₆₀₀) to account for growth effects.
- Curve Fitting: Plot the normalized output against the ligand concentration and fit the data to a Hill equation model [4]:
Output = Minimum + (Maximum - Minimum) * [Ligand]^nH / (K^nH + [Ligand]^nH)where:Minimumis the basal signal level.Maximumis the saturated signal level.Kis the ligand concentration at half-maximal output (an indicator of affinity).nHis the Hill coefficient, describing cooperativity.
Quantification from the Dose-Response Curve
After curve fitting, the key metrics are calculated as follows:
- Dynamic Range:
Maximum / Minimum[15]. - Sensitivity: The
Kvalue and thenH(Hill coefficient) together describe the sensitivity and sharpness of the response [15]. - Specificity: Determine the dynamic range for the target ligand and several structurally similar molecules. A biosensor with high specificity will show a significantly higher dynamic range for its intended target [15].
Engineering Strategies for Performance Enhancement
Native TFBs often require optimization for practical application. The following strategies enable fine-tuning of sensitivity, dynamic range, and specificity.
Table 2: Engineering Strategies for Optimizing TFB Performance
| Target Metric | Engineering Strategy | Specific Method | Mechanism of Action |
|---|---|---|---|
| Sensitivity & Dynamic Range | Promoter Engineering [15] | Vary the strength of the promoter controlling the reporter gene or the TF itself. | Alters the number of output molecules or TF abundance, directly affecting signal amplitude and background noise. |
| RBS Tuning [4] [15] | Modify the Ribosome Binding Site (RBS) sequence upstream of the TF or reporter gene. | Fine-tunes the translation efficiency, controlling the intracellular concentration of the TF and the reporter protein. | |
| Operator Site Modification [15] | Mutate the transcription factor binding site (TFBS or operator) on the promoter. | Alters the binding affinity of the TF for the DNA, which changes the basal expression level and induction threshold. | |
| Plasmid Copy Number [4] | Use plasmids with different origins of replication. | Varies the gene dosage of the biosensor components, impacting the absolute number of sensors per cell. | |
| Specificity | TF Ligand-Binding Domain Engineering [1] [15] [16] | Use directed evolution or rational design to mutate residues in the TF's ligand-binding pocket. | Reshapes the binding pocket to sterically hinder non-target ligands or create new favorable interactions with the target. |
| Chimeric TF Construction [3] | Fuse the DNA-binding domain of one TF with the ligand-binding domain of another. | Creates novel biosensors with hybrid functions, combining the DNA recognition of one TF with the sensing capability of another. |
Advanced high-throughput platforms like Sensor-seq are revolutionizing TFB engineering. This method uses RNA barcoding and deep sequencing to simultaneously quantify the performance (F-score) of thousands of TtgR protein variants in response to various ligands, efficiently identifying rare variants with desired specificity and dynamic range [16].
The Scientist's Toolkit: Essential Research Reagents
Developing and applying high-performance TFBs relies on a suite of core reagents and methodologies.
Table 3: Essential Reagents and Tools for TFB Research
| Reagent / Tool Category | Specific Examples | Function in Biosensor R&D |
|---|---|---|
| Model Transcription Factors | TetR, AraC, TrpR, TtgR, LuxR [10] [16] | Well-characterized scaffolds for engineering new biosensors; their structural and functional data provide a starting point for design. |
| Reporter Genes | GFP (Green Fluorescent Protein), RFP (Red Fluorescent Protein), Luciferase, Enzymatic Reporters (e.g., LacZ) [1] [10] | Generate a quantifiable output signal (optical, luminescent, colorimetric) that is linked to ligand concentration. |
| High-Throughput Screening Platforms | Flow Cytometry, Microfluidics, Sensor-seq [1] [16] | Enable the sorting and analysis of vast libraries of microbial variants to isolate those with superior biosensor performance. |
| Computational & AI Tools | Cello (Genetic Circuit Design), DeepTFactor (TF Prediction), AlphaFold (Protein Structure Prediction) [1] [10] | Facilitate in silico design, optimization, and prediction of biosensor components and their interactions before experimental testing. |
| Directed Evolution Techniques | Error-prone PCR, Site-saturation mutagenesis [15] [16] | Create diverse libraries of TF mutants to evolve novel ligand specificity, sensitivity, and improved dynamic range. |
| Fen1-IN-6 | Fen1-IN-6, MF:C12H8N2O5S2, MW:324.3 g/mol | Chemical Reagent |
| Wee1-IN-6 | Wee1-IN-6, MF:C45H52FN11O4, MW:830.0 g/mol | Chemical Reagent |
Application in Dynamic Regulation and Screening
Optimizing these metrics is not an academic exercise; it directly enables advanced applications in metabolic engineering.
- High-Throughput Screening (HTS): A large dynamic range is critical for efficiently separating high-producing cells from low-producers in large libraries [1] [4]. For example, TFBs for biofuels like butanol or fatty acid-derived products have been used to screen strain libraries, identifying mutants with significantly improved titers [4].
- Dynamic Metabolic Regulation: Biosensors can be designed to automatically downregulate competitive pathways or upregulate bottleneck enzymes when a key intermediate accumulates [3] [4]. The sensitivity and response time of the biosensor determine how quickly and effectively the metabolic flux can be rebalanced, leading to increased product yields and robustness in scaled-up fermentations [3] [15].
In conclusion, a deep understanding and systematic optimization of sensitivity, dynamic range, and specificity are fundamental to harnessing the full potential of transcription factor-based biosensors. Through strategic engineering and the use of advanced screening and computational tools, researchers can develop precision tools that drive innovation in drug development, bio-based chemical production, and fundamental biological research.
The journey to understanding and engineering cellular control began with the seminal study of the lac operon in E. coli. This system provided the first model for inducible gene regulation, demonstrating how a transcription factor (LacI) could repress a set of genes (lacZYA) in the absence of its inducer (allolactose or the synthetic analog IPTG). Upon inducer binding, LacI undergoes a conformational change, dissociates from the operator site, and allows transcription to proceed [17]. This fundamental "on/off" switch mechanism, governed by the interaction between a transcription factor (TF), a small molecule, and DNA, established the core principle upon which the entire field of synthetic biology would later build. The quest to move from observing this natural system to deliberately engineering it for predefined purposes forms the historical arc that has led to the development of sophisticated transcription factor-based biosensors (TFBs) for dynamic regulation. These biosensors are genetically encoded devices that use transcription factors to convert the intracellular concentration of a specific metabolite into a measurable output, enabling real-time monitoring and control of biological processes [9] [4].
From Natural System to Engineering Framework
The lac operon is more than a biological concept; it is a functional module that can be extracted and repurposed. Synthetic biology, inspired by engineering disciplines, sought to standardize and simplify such biological parts to make them predictable and reliable [18]. The lac system's components—the LacI repressor, its operator sequence, and the inducible promoter—became the first standardized parts in the synthetic biology toolkit.
Early engineering efforts revealed the system's complexity. A theoretical model of a simple regulatory region with three activator binding sites requires accounting for at least eight possible states and nine unique molecular parameters [18]. To make this complexity manageable, synthetic biologists adopted a reductionist approach, systematically simplifying systems to distill them to their essential features. This philosophy of "bending nature to understand it" enabled a predictive understanding of biological processes [18].
Table 1: Evolution from Natural Operon to Engineered Biosensor
| Feature | Natural Lac Operon | Modern Engineered TFB |
|---|---|---|
| Core Principle | Negative inducible genetic switch | Programmable molecular sensor-actuator |
| Primary Function | Metabolic adaptation to carbon source | Dynamic pathway control, high-throughput screening, metabolite detection |
| Key Components | LacI, allolactose, native pLac promoter | Engineered TFs, novel ligands, synthetic promoters & RBSs |
| Typical Output | Metabolic enzymes for lactose digestion | Fluorescent proteins, enzymes for colorimetric change, survival markers |
| Tunability | Limited by natural evolution | Highly tunable via promoter strength, RBS engineering, and TF engineering |
| Application Scope | Single-organism metabolism | Cross-species, cell-free systems, multi-input logic gates |
The conceptual leap from the natural operon to an engineered biosensor is summarized in Table 1. The modern TFB is a modular device comprising a sensing component (the transcription factor) and a reporting component (a promoter controlling a reporter gene) [9]. The fundamental operational logic, however, remains unchanged from the lac paradigm: a small molecule binds to a TF, inducing a conformational change that alters its DNA-binding affinity, thereby regulating downstream gene expression [8].
Figure 1: Core Mechanism of a Transcription Factor-Based Biosensor. The binding of a specific inducer molecule causes a conformational change in the transcription factor, altering its ability to bind DNA and regulate the expression of a reporter gene.
Engineering and Optimizing Biosensor Performance
Wild-type transcription factors like LacI rarely possess the optimal characteristics for applied biosensing. A primary challenge in the field is the limited number of known metabolite-activated TFs compared to the vast number of compounds amenable to biomanufacturing [9] [10]. Consequently, significant research focuses on engineering and optimizing TFBs to enhance their performance for specific tasks. Key performance metrics include:
- Dynamic Range: The fold difference between the fully induced ("ON") and uninduced ("OFF") output states [4] [8].
- Sensitivity: The lowest concentration of ligand that elicits a detectable response, often defined by the EC50 (half-maximal effective concentration) [4].
- Specificity: The ability to distinguish the target ligand from structurally similar molecules [4].
- Operating Range: The concentration window over which the biosensor response is linear [9].
Table 2: Key Engineering Strategies for Optimizing Transcription Factor-Based Biosensors
| Engineering Target | Strategy | Purpose & Outcome |
|---|---|---|
| Transcription Factor (TF) | Directed evolution & protein engineering [9] [16] | Alter ligand specificity, improve sensitivity, or reduce cross-talk. |
| TF Structure | Rational design of ligand-binding domain [4] | Modulate affinity for the target ligand and allosteric control. |
| Promoter & Operator | Mutagenesis of TF binding site (operator) [8] | Fine-tune the binding affinity of the TF, altering leakiness and dynamic range. |
| Ribosome Binding Site (RBS) | Library-based screening of RBS variants [4] | Optimize translation efficiency of the reporter protein to maximize output signal. |
| Genetic Context | Modulation of plasmid copy number [4] | Balance gene dosage effects to prevent cellular burden and maintain sensitivity. |
| Circuit Architecture | Incorporation of genetic amplifiers [4] | Amplify the output signal, thereby increasing the dynamic range and sensitivity. |
Engineering a TFB requires a holistic approach, as summarized in Table 2. For instance, the TF itself can be engineered through directed evolution. A groundbreaking approach, Sensor-seq, enables highly multiplexed design of allosteric transcription factors for new ligands. This method involves creating vast libraries of TF variants (e.g., 17,737 variants of TtgR) and using RNA barcoding and deep sequencing to quantitatively measure the ligand-induced response (F-score) of each variant in a high-throughput manner [16]. This platform has successfully created biosensors for non-native ligands like the opiate analog naltrexone and the antimalarial drug quinine [16].
Experimental Workflow for Biosensor Development and Validation
The development of a functional biosensor follows a structured pipeline from part identification to performance characterization. The workflow can be broken down into several key stages, as illustrated below and detailed in the accompanying protocol.
Figure 2: Generalized Workflow for Biosensor Construction and Testing.
Protocol: Construction and Characterization of a Phenolic Acid Biosensor
This protocol is adapted from a study that developed biosensors for eleven different phenolic acids [13].
1. Part Identification & Library Design:
- Objective: Identify a transcription factor and its cognate promoter that respond to a target molecule (e.g., a phenolic acid).
- Method:
- Perform a multi-genome bioinformatic screen to identify potential TF-promoter pairs. Key databases include RegulonDB (for E. coli), RegPrecise (for prokaryotes), and P2TF [9] [13].
- Design a genetic library if engineering is required. For a novel ligand, use a platform like Sensor-seq [16]: select a promiscuous aTF scaffold (e.g., TtgR), generate a library of sequence variants via phylogeny-guided diversification, and clone them into a screening construct that links each variant to a unique RNA barcode.
2. Genetic Circuit Construction:
- Objective: Assemble the biosensor circuit in an appropriate plasmid vector.
- Method:
- Clone the identified inducible promoter upstream of a reporter gene (e.g., for fluorescent protein like RFP or GFP) [13].
- Ensure the gene for the cognate transcription factor is also present in the system, typically expressed constitutively from a separate promoter.
- The final plasmid is the "screening construct" used for transformation.
3. Host Transformation & Cultivation:
- Objective: Introduce the biosensor construct into the host chassis.
- Method:
- Transform the plasmid into model bacterial hosts such as Escherichia coli, Cupriavidus necator, or Pseudomonas putida [13].
- Plate transformed cells on selective media and incubate to obtain single colonies.
- Inoculate liquid cultures from single colonies and grow to the desired cell density (typically mid-log phase) under appropriate conditions.
4. Induction Assay & High-Throughput Screening:
- Objective: Measure the biosensor's response to a range of ligand concentrations.
- Method:
- For a characterized biosensor: Distribute cell cultures into a multi-well plate. Add a gradient of the target ligand (e.g., phenolic acids, naltrexone, quinine) to different wells, including a no-ligand negative control. Incubate for a defined period to allow gene expression [13].
- For a large variant library (Sensor-seq): Pool the entire library of clones. Split the culture and dose with either the target ligand or a vehicle control. Harvest samples during log-phase growth for RNA extraction and plasmid DNA isolation [16].
5. Data Acquisition & Analysis:
- Objective: Quantify the output and calculate performance metrics.
- Method:
- For fluorescent reporters: Use a plate reader to measure fluorescence intensity. Normalize fluorescence to cell density (OD600) [13].
- For Sensor-seq: Prepare cDNA from total RNA and sequence it along with the plasmid DNA library. Map each TF variant to its output via the unique barcode. Calculate an F-score for each variant: the normalized ratio of reporter transcript levels in the presence vs. absence of ligand [16].
- Plot dose-response curves (output vs. ligand concentration) and fit with the Hill equation to determine key parameters like dynamic range, EC50, and Hill coefficient [4].
6. Iterative Optimization:
- Objective: Improve biosensor performance based on initial characterization data.
- Method:
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagent Solutions for TFB Development
| Reagent / Material | Function in Research | Example & Context |
|---|---|---|
| Model Chassis | Host organism for biosensor construction and testing. | E. coli BL21, Pseudomonas putida, Cupriavidus necator; chosen for genetic tractability and application context [13]. |
| Reporter Genes | Provides a measurable output signal linked to TF activity. | Fluorescent Proteins (GFP, RFP), Enzymes (XylE, LacZ), RNA Aptamers (Mango III) [13] [17]. |
| Database Resources | In silico source for identifying native TF-promoter pairs. | RegulonDB, RegPrecise, P2TF, JASPAR [9]. |
| Screening Platform | High-throughput method for identifying functional biosensors from large variant libraries. | Sensor-seq: uses RNA barcoding and deep sequencing [16]. FACS: for screening based on fluorescence [19]. |
| Cell-Free Systems | Simplified, rapid prototyping environment bypassing cell walls and complex physiology. | Freeze-dried crude extracts (e.g., from E. coli); enable biosensing by just adding water, DNA, and the ligand [14] [17]. |
| Directed Evolution Tools | Methods to generate genetic diversity in TF sequences for new functions. | Error-prone PCR, Site-saturation mutagenesis, DNA shuffling [9] [16]. |
| Vem-L-Cy5 | Vem-L-Cy5, MF:C63H68F5N7O9S, MW:1194.3 g/mol | Chemical Reagent |
| Liproxstatin-1-15N | Liproxstatin-1-15N|Ferroptosis Inhibitor | Liproxstatin-1-15N is a potent N-15 labeled ferroptosis inhibitor for research. This product is for Research Use Only (RUO) and is not intended for diagnostic or therapeutic use. |
Applications in Metabolic Engineering and Synthetic Biology
The transition of TFBs from research tools to core components in engineering biology is most evident in their applications. A primary use case is in high-throughput screening (HTS) for metabolic engineering. TFBs can be used to screen vast libraries of engineered microbes to identify rare, high-producing variants. By linking the production of a desired metabolite (e.g., an advanced biofuel) to the expression of a fluorescent reporter, fluorescence-activated cell sorting (FACS) can be used to isolate top producers from a population of millions of cells [4] [19].
Beyond screening, TFBs enable dynamic metabolic regulation. In this approach, a biosensor detecting an intermediate metabolite automatically upregulates or downregulates key pathway enzymes to balance metabolic flux. This creates a self-regulating cell factory that optimizes production in real-time, overcoming the limitations of static engineering strategies [9] [4]. For example, a TFB for a key intermediate can be designed to downregulate a competing pathway, thereby redirecting carbon flux toward the desired product [4].
Furthermore, the utility of TFBs extends into environmental and clinical monitoring. Engineered biosensors for specific molecules, such as the opiate naltrexone or the antimalarial quinine, can be deployed in cell-free systems for diagnostic purposes [16]. These systems, often lyophilized for stability, function as low-cost, field-deployable sensors for contaminants or health biomarkers [17].
The field of transcription factor-based biosensors is poised for transformative growth, driven by several emerging trends. The integration of artificial intelligence (AI) and machine learning is accelerating the design process. Tools like DeepTFactor predict new transcription factors from sequence data, while platforms like Sensor-seq generate large datasets ideal for training models to predict the functional outcomes of specific mutations, moving the field toward predictive biosensor design [14] [9] [10].
There is also a strong push toward applying these tools in non-model organisms and complex consortia, which may be more robust for industrial bioprocesses [14]. Finally, the use of cell-free systems is democratizing access to synthetic biology and enabling the creation of portable, shelf-stable biosensors for applications ranging from education to point-of-care diagnostics [14] [17].
In conclusion, the path from the Lac operon to modern synthetic biology represents a profound shift from observation to mastery. The simple switch discovered in E. coli has been deconstructed, understood, and rebuilt into a versatile engineering paradigm. Today's TFBs are sophisticated devices capable of sensing, computing, and actuating within living cells, enabling unprecedented control over biological systems for manufacturing, medicine, and environmental sustainability. As our ability to design and optimize these biological tools continues to improve, their impact on science and technology is bound to deepen, firmly establishing synthetic biology as a predictive engineering discipline.
Engineering and Deploying Biosensors: From Laboratory Design to Real-World Solutions
The engineering of genetic circuits represents a cornerstone of synthetic biology, enabling the programming of living cells to perform complex functions, from bio-based chemical production to advanced therapeutic applications [20]. These circuits require the precise integration of core components—promoters, operators, and reporter genes—to process biological information and generate measurable outputs. Within the broader research context of transcription factor-based biosensors for dynamic regulation, the construction of reliable genetic circuits is paramount [10]. Such biosensors, which often utilize allosteric transcription factors (aTFs) to detect specific metabolites and elicit a programmed response, depend on well-characterized genetic parts to function predictably inside living cells [10]. This guide provides an in-depth technical overview of the fundamental principles and methodologies for constructing these circuits, framing them as essential enabling technologies for the development of sophisticated biosensing and regulatory systems.
Core Components of Genetic Circuits
Promoters
Promoters are DNA sequences where RNA polymerase binds to initiate transcription. In synthetic biology, both constitutive and inducible promoters are used. Inducible promoters are particularly valuable for biosensor applications, as their activity can be regulated by specific transcription factors that, in turn, respond to effector molecules [20] [10]. The strength of a promoter, determined by its -35 and -10 box sequences, dictates the baseline and maximum levels of gene expression, which must be carefully balanced for a circuit to function as intended [20].
Operators
Operators are short DNA sequences that serve as binding sites for transcription factor (TF) proteins. When a TF binds to its operator, it can either activate or repress transcription from an adjacent promoter. In the context of biosensors, allosteric transcription factors (aTFs) undergo a conformational change upon binding a specific ligand (e.g., a small molecule metabolite), which alters their ability to bind DNA and thereby regulates gene expression [10]. The precise number, type, and arrangement of operator sites in a regulatory region is a key determinant of the logical operation (e.g., AND, NOT) performed by the circuit [21].
Reporter Genes
Reporter genes produce a measurable output that correlates with the activity of a genetic circuit. The Green Fluorescent Protein (GFP) is a widely used reporter that allows for quantitative measurement of gene expression using fluorometry [21]. The choice of reporter is critical, as its degradation rate and expression level can impact the ability to accurately measure circuit dynamics [20].
Table 1: Core Components of a Genetic Circuit
| Component | Function | Key Characteristics | Example |
|---|---|---|---|
| Promoter | Initiates transcription by recruiting RNA polymerase. | Defined by -35/-10 sequences; can be constitutive or inducible. | Placq (constitutive), luxpR (inducible) [21] |
| Operator | Protein-binding site for transcriptional regulation. | Specific DNA sequence recognized by a transcription factor. | LacI operator, LuxR activator binding site [21] |
| Reporter Gene | Encodes a measurable output for monitoring circuit activity. | Must produce a quantifiable signal (e.g., fluorescence). | Green Fluorescent Protein (gfp) [21] |
Figure 1: Information flow in a TF-based biosensor. An effector molecule binds a transcription factor, which then interacts with an operator to regulate promoter activity and reporter gene output.
Genetic Circuit Design and Function
The design of genetic circuits involves the strategic interconnection of promoters, operators, and coding sequences to create a desired logical function. A foundational goal is to establish a common signal carrier, such as RNA polymerase flux controlled by promoters, which simplifies the connection of smaller circuits into larger, more sophisticated systems [20]. Circuit dynamics are profoundly influenced by the chosen regulatory molecules, which can include DNA-binding proteins, invertases, and CRISPR-based systems [20].
Regulator Classes for Circuit Design
- DNA-Binding Proteins: These are the most common regulators, including repressors like LacI and TetR, and activators. They function by binding to specific operator sequences to either block (repress) or recruit (activate) RNA polymerase [20]. Simple NOT and NOR logic gates can be built using repressible promoters, while AND gates have been constructed using activators that require chaperones or split proteins [20].
- CRISPRi/a: Catalytically inactive Cas9 (dCas9) can be fused to repressor or activator domains and targeted to specific DNA sequences via guide RNAs. This system offers high designability, as the targeting is determined by an RNA sequence, allowing for a large set of orthogonal regulators [20].
- Invertases: These are site-specific recombinases that permanently invert DNA segments between binding sites. They are ideal for building memory circuits, as the flipped state is maintained without continuous energy input [20].
Construction of a Genetic AND Gate: A Case Study
This section details a specific experimental methodology for constructing a genetic AND gate, demonstrating the assembly of core components to create a Boolean logic function within a cell [21].
Experimental Protocol
1. Plasmid Design and Standardization:
- A reporter plasmid (
pSB-GFP) is used, which contains a promoterlessgfpgene. - The regulatory region upstream of
gfpis standardized to include an upstream multi-cloning site (for promoter parts) and a downstream standard region designed for the sequential insertion of operator parts using specific restriction enzymes (BglIIandMluI).
2. Assembly of Operator Parts:
- An operator part is synthesized as a DNA module flanked by
BamHIandMluIrestriction sites. TheBamHIsticky end is compatible with theBglIIsite on the plasmid. - The operator part is ligated into the
BglII/MluI-digested plasmid. This ligation destroys the originalBglIIsite but leaves a newBglIIsite on the inserted part, allowing for the repeated insertion of additional operator parts using the same procedure.
3. Circuit Construction:
- A constitutive promoter part (
Placq) is first inserted into the upstream multi-cloning site to createPlacq-GFP. - To build a LacI-repressed gate, a single
LacIoperator part is inserted into the downstream standard region, creatingPlacq-LacI-GFP. A version with two operators (Placq-LacItandem-GFP) is also constructed for stronger repression. - To build a LuxR-activated gate, a
LuxRoperator part (containing aluxpRpromoter, a LuxR-binding site, and aLuxRexpression cassette) is assembled. - Finally, the AND gate (
PLuxR-LacItandem-GFP) is constructed by inserting the twoLacIoperator parts into the downstream standard region of thePLuxR-GFPplasmid.
4. Reporter Assay and Validation:
- Each constructed plasmid is transformed into an E. coli strain expressing
LacI(e.g.,DH5αLacI). - Transformed cells are grown in culture with different combinations of inducers: Isopropyl β-D-1-thiogalactopyranoside (IPTG) for LacI and N-(β-ketocaproyl)-DL-homoserine lactone (AHL) for LuxR.
- After a suitable incubation period, GFP fluorescence is measured using a fluorometer. Fluorescence intensity is normalized to cell density.
- A functional AND gate will show high fluorescence output only in the presence of both IPTG and AHL.
Table 2: Key Research Reagents and Materials
| Reagent/Material | Function in Experiment |
|---|---|
| pSB-GFP Plasmid | Backbone vector with promoterless GFP reporter gene [21]. |
| Operator Parts | Standardized DNA modules containing protein-binding sites (e.g., for LacI, LuxR) [21]. |
| Restriction Enzymes | Molecular tools for digesting DNA at specific sequences for assembly (e.g., BglII, MluI, BamHI) [21]. |
| E. coli DH5αLacI | Host cell strain that provides the LacI repressor protein for inducible system function [21]. |
| Inducers (IPTG, AHL) | Small molecules that trigger the biosensor by binding to and altering their respective transcription factors [21]. |
| Fluorometer | Instrument for quantifying the output signal (GFP fluorescence) from the genetic circuit [21]. |
Figure 2: Genetic AND gate logic. Only when AHL activates LuxR AND IPTG inactivates LacI repression does transcription of GFP occur.
Expected Results and Analysis
In the case study [21], the constructed AND gate (PLuxR-LacItandem-GFP) exhibited the following behavior:
- No Inducers: Background fluorescence.
- IPTG Only: Low fluorescence (LacI repression is lifted, but LuxR is not activated).
- AHL Only: Low fluorescence (LuxR is activated, but LacI repression blocks transcription).
- IPTG and AHL: High fluorescence (5.7-fold increase over background), as both repression is lifted and activation occurs.
This result confirms the successful integration of two biological inputs into a single transcriptional output via the modular assembly of standardized operator parts.
Advanced Applications in Biosensing and Dynamic Regulation
The construction of basic logic gates provides the foundation for developing advanced transcription factor-based biosensors. These biosensors are crucial tools for two primary applications in synthetic biology: screening and dynamic regulation [10].
Screening Production Strains: Biosensors can be designed to detect and respond to the intracellular accumulation of a desired product, such as a bio-based chemical. A biosensor circuit can be linked to a reporter gene like GFP, allowing researchers to use fluorescence-activated cell sorting (FACS) to identify and isolate high-producing microbial strains from a library [10].
Dynamic Regulation of Metabolic Pathways: Biosensors enable dynamic control strategies within engineered cells. Instead of constitutively expressing all enzymes in a metabolic pathway, a biosensor can detect an intermediate metabolite's buildup. This signal can then trigger the expression of downstream enzymes, automatically balancing metabolic flux to maximize product yield and minimize the accumulation of toxic intermediates [10].
The reliability of these advanced applications hinges on the precise construction and predictable performance of the underlying genetic circuits, underscoring the necessity for robust design principles, well-characterized parts, and standardized assembly methods [20] [10].
The engineering of biological systems for applications in therapeutics, diagnostics, and bioproduction relies heavily on the precise control of gene expression. Within the specific context of transcription factor (TF)-based biosensors, dynamic regulation has emerged as a critical capability for advancing metabolic engineering and synthetic biology [14] [10]. These biosensors, which convert the presence of a target metabolite into a measurable genetic output, enable real-time monitoring and control of biosynthetic pathways, thereby overcoming limitations in natural product yield [14]. The performance and evolutionary longevity of these systems are governed by two fundamental layers of control: transcriptional and translational.
Transcriptional control operates at the level of RNA synthesis, where transcription factors and promoters regulate when and how frequently a gene is transcribed. Translational control, in contrast, acts at the protein synthesis level, fine-tuning the efficiency with which mRNA is translated into functional protein, often through mechanisms involving ribosome binding sites, small RNAs, and upstream open reading frames (uORFs) [22]. While transcriptional control provides the primary on/off switch for gene expression, translational regulation offers a faster, more direct means to modulate protein output, which is crucial for rapid cellular adaptation [22]. This guide provides an in-depth examination of the core strategies for tuning both transcriptional and translational processes, with a specific focus on their application in TF-based biosensors for dynamic regulation research.
Transcriptional Control Strategies
Transcriptional control forms the foundational layer of genetic regulation in engineered biosensors. It encompasses the suite of mechanisms that regulate the initiation and rate of transcription, primarily through the interaction of transcription factors with specific DNA sequences.
Allosteric Transcription Factors (aTFs) and Their Engineering
Allosteric transcription factors (aTFs) are proteins that undergo conformational changes upon binding to specific small molecules (ligands), which in turn alters their affinity for operator DNA sequences and modulates downstream gene expression [10]. This inherent switching mechanism makes them ideal sensing components for synthetic genetic circuits. The design space for aTFs includes several modes of action: repression of activator aTF, activation of repressor aTF, repression of repressor aTF, or activation of activator aTF [10].
A significant challenge in biosensor engineering is the limited number of natural aTFs relative to the vast array of compounds targeted for detection. To address this constraint, several advanced engineering strategies have been developed:
- Sensor-seq Platform: This high-throughput method designs aTFs to sense non-native ligands. It involves creating vast libraries of aTF variants (e.g., 17,737 variants of TtgR) and screening them against target ligands using RNA barcoding and deep sequencing to quantify ligand-induced responses (F-score) [16]. This platform successfully created biosensors for diverse ligands, including the opiate analog naltrexone and the antimalarial drug quinine.
- Structure-Guided Design: Computational tools, including molecular dynamics (MD) simulations, guide the rational design of artificial TFs. For instance, a progesterone biosensor was designed by fine-tuning the linker between the DNA-binding domain (QFDBD) and activation domain (QFAD) to achieve the desired conformational change upon ligand binding [23].
- Homology and AI-Based Prediction: Bioinformatics approaches leverage known transcriptional regulator families (e.g., LysR, TetR, AraC) to discover new aTFs through sequence homology in genomic and metagenomic databases [10]. Furthermore, artificial intelligence tools like DeepTFactor predict novel TFs from sequence data, expanding the discoverable space of potential biosensors [10].
Promoter Engineering and Synthetic Promoters
The promoter region is a critical determinant of transcriptional activity and specificity. Engineering these regions allows for fine-tuning of biosensor performance characteristics, such as dynamic range, leakiness, and host compatibility.
- Synthetic Promoter Design: The creation of entirely synthetic promoters, such as the promoter QT (composed of QUAS, a 10-bp spacer, and a T7 promoter), enables precise control over transcription initiation by designed artificial TFs [23].
- Operator Sequence Modification: Altering the operator sequences to which aTFs bind can significantly impact the binding affinity of the TF and, consequently, the repression or activation strength of the promoter. This is a key parameter for tuning the transfer function of the biosensor.
- Library-Based Screening: Generating diversified promoter libraries through mutagenesis of core promoter elements allows for the experimental screening and selection of variants with desired expression levels and induction profiles.
Table 1: Key Quantitative Metrics for Evaluating Transcriptional Biosensor Performance
| Metric | Description | Typical Experimental Measurement |
|---|---|---|
| Dynamic Range | The ratio of output signal in the presence of a saturating ligand concentration to the output signal in its absence (basal expression). | Flow cytometry, microplate reader (e.g., GFP fluorescence). |
| ECâ‚…â‚€ / ICâ‚…â‚€ | The effective concentration of ligand that induces 50% of the maximum activation or repression. | Dose-response curve with ligand titration. |
| Sensitivity (LOD) | The lowest concentration of ligand that produces a detectable output signal significantly above the background noise. | Limit of Detection (LOD) calculation from dose-response data. |
| F-score | A normalized ratio of reporter transcript levels with and without ligand, used in high-throughput screens like Sensor-seq [16]. | RNA sequencing (RNA-seq). |
| Half-life (Ï„â‚…â‚€) | The time taken for the population-level functional output of a circuit to fall by 50% due to evolutionary degradation [24]. | Long-term growth and measurement assays (e.g., over serial passaging). |
Experimental Protocol: Sensor-seq for High-Throughput aTF Screening
The following protocol outlines the key steps for implementing the Sensor-seq platform to identify aTF variants responsive to new ligands [16].
- Library Construction: Clone a diversified library of aTF variants (e.g., via site-saturation mutagenesis) into a screening plasmid. Each variant is placed in cis with a randomized 16-nucleotide barcode, separated by a constant region containing the aTF's native promoter driving a reporter transcript.
- Ligand Exposure and Cell Harvesting: Transform the pooled plasmid library into the host organism (e.g., E. coli). Grow separate cultures dosed with the target ligand and a vehicle control. Harvest cells during the log-phase growth for both total RNA and plasmid DNA extraction.
- RNA Sequencing and Mapping Genotype to Phenotype:
- RNA-seq: Prepare cDNA from the total RNA and sequence it to quantify the abundance of each reporter barcode transcript, which reflects the activity of the corresponding aTF variant under the given condition.
- Genotype-Barcode Linkage: Use PCR and Golden Gate Assembly to physically link each aTF variant sequence in the plasmid DNA to its associated barcode. Sequence these constructs to create a mapping between every variant and its barcode(s).
- Data Analysis and F-score Calculation: For each variant, calculate the F-score using the formula:
F-score = (Normalized cDNA count with ligand) / (Normalized cDNA count without ligand)Normalization is performed using the plasmid DNA counts for each barcode. Variants with an F-score significantly greater than 1 are considered functional biosensors for the target ligand.
Translational Control Strategies
While transcriptional control initiates gene expression, translational regulation provides a faster, more direct layer of tuning that determines the final protein output from an mRNA transcript. This post-transcriptional control is crucial for rapid adaptive responses and for minimizing the metabolic burden associated with transcription, thereby enhancing the evolutionary stability of genetic circuits [24] [22].
Mechanisms of Translational Regulation
Translational regulation fine-tunes protein synthesis at several key stages:
- Initiation Control: This is the most critical rate-limiting step. Efficiency is primarily governed by the affinity of the ribosome binding site (RBS) for the ribosome, the secondary structure of the mRNA around the start codon, and the availability of initiation factors [22]. Engineering the RBS sequence is a primary method for tuning translation initiation and, consequently, protein expression levels.
- Small RNA (sRNA) Mediated Control: Small RNAs can act as post-transcriptional regulators by base-pairing with target mRNAs, which can block ribosome access or promote mRNA degradation. In synthetic biology, sRNAs are exploited in post-transcriptional controllers that silence circuit RNA. These controllers have been shown to outperform transcriptional controllers in extending the evolutionary longevity of gene circuits because they provide strong control with reduced burden [24].
- Upstream Open Reading Frames (uORFs): uORFs are short coding sequences located in the 5' untranslated region (5' UTR) of an mRNA. The translation of a uORF can regulate the translation of the downstream main ORF by modulating ribosome progression and re-initiation. uORFs are a natural mechanism for fine-tuning gene expression in response to cellular conditions, and their manipulation (e.g., using antisense oligonucleotides to block the uORF start codon) is an emerging translational control strategy [22].
- Elongation and Termination: The speed of ribosomal elongation and the efficiency of termination can also impact overall protein yield and co-translational folding. While more complex to engineer, these processes represent additional points of potential control.
Experimental Protocol: Ribosome Profiling (Ribo-Seq) for Translational Efficiency Analysis
Ribosome profiling is a powerful method that provides a genome-wide, nucleotide-resolution snapshot of translation by sequencing ribosome-protected mRNA fragments [22].
- Cell Lysis and Nuclease Digestion: Rapidly lyse cells to freeze translational states. Treat the lysate with RNase I, which degrades mRNA regions not protected by bound ribosomes.
- Ribosome-Protected Fragment (RPF) Purification: Isolate the ribosome-protected mRNA fragments (typically ~30 nucleotides) by size selection through sucrose cushion centrifugation or gel extraction.
- Library Preparation and Sequencing: Convert the purified RPFs into a sequencing library. In parallel, prepare a standard RNA-seq library from the same sample's total mRNA.
- Data Analysis and Translational Efficiency (TE) Calculation:
- Map the RPF sequencing reads and the RNA-seq reads to a reference genome.
- Quantify the ribosome density for each gene from the RPF data.
- Quantify the mRNA abundance for each gene from the RNA-seq data.
- Calculate the Translational Efficiency (TE) for each gene using the formula:
TE = (Normalized RPF read count) / (Normalized RNA-seq read count) - Changes in TE (ΔTE) under different conditions (e.g., with/without ligand, or in a disease state) reveal genes that are subject to specific translational regulation.
Table 2: Core Methodologies for Studying Translational Regulation
| Method | Principle | Key Output | Advantages |
|---|---|---|---|
| SUnSET Assay | Incorporation of puromycin, a tRNA analog, into nascent peptides during active translation, detected via immunoblotting. | Global rate of protein synthesis. | Simple, measures translation in whole cells/tissues. |
| Ribosome Profiling (Ribo-Seq) | Deep sequencing of ribosome-protected mRNA fragments (RPFs). | Genome-wide map of ribosome positions and density. | Nucleotide resolution; identifies actively translated ORFs (including uORFs); calculates TE. |
| Polysome Profiling | Sucrose gradient centrifugation to separate mRNAs based on the number of bound ribosomes. | Transcriptome-wide assessment of translation activity. | Distinguishes highly translated from poorly translated mRNAs. |
| TRAP / Ribo-tag | Immunoprecipitation of epitope-tagged ribosomes from specific cell types in a complex tissue. | Translatome of a specific cell population. | Cell-type-specific translational information in vivo. |
Integration and Application in Dynamic Regulation
The true power of transcriptional and translational control is realized when they are intelligently integrated to create robust, dynamic regulatory systems for TF-based biosensors.
Dynamic Control in Metabolic Engineering
TF-based biosensors enable two primary application paradigms in metabolic engineering:
- High-Throughput Screening: Biosensors can be designed to link the production of a desired metabolite to a easily measurable output (e.g., fluorescence). This allows for the rapid screening of vast mutant libraries to identify high-producing strains, dramatically accelerating the strain optimization process [14] [10].
- Closed-Loop Dynamic Regulation: In this advanced strategy, a biosensor continuously monitors the concentration of a key pathway intermediate. The output of the biosensor is then linked to a genetic controller that dynamically regulates the expression of pathway enzymes. This allows the cell to autonomously balance metabolic flux, avoid the accumulation of toxic intermediates, and maximize product yield without researcher intervention [14] [10]. For example, a TF that senses a fatty acid intermediate could dynamically control the expression of upstream enzymes in the biofuel synthesis pathway.
Enhancing Evolutionary Longevity with Genetic Controllers
A major challenge in synthetic biology is the evolutionary degradation of engineered gene circuits due to mutations that reduce metabolic burden [24]. "Host-aware" computational frameworks have been used to design genetic controllers that extend circuit functional half-life.
- Negative Autoregulation: This transcriptional feedback topology, where a TF represses its own promoter, can reduce expression noise and burden, prolonging short-term performance [24].
- Growth-Based Feedback: Controllers that couple circuit output to host growth rate can significantly extend the functional half-life (Ï„â‚…â‚€) of a circuit. Simulations show that growth-based feedback can outperform intra-circuit feedback in the long term [24].
- Multi-Input Controllers: The most effective designs combine multiple control inputs (e.g., circuit output and growth rate) and actuation methods (transcriptional and post-transcriptional). These complex controllers can improve circuit half-life over threefold without needing to couple to an essential gene [24].
The Scientist's Toolkit: Essential Research Reagents and Solutions
Table 3: Key Reagents and Tools for Transcriptional and Translational Control Research
| Reagent / Tool | Function | Example Use Case |
|---|---|---|
| Allosteric Transcription Factor (aTF) Scaffolds | The core sensing component of a biosensor. Can be engineered for new ligands. | TtgR, a promiscuous multidrug efflux regulator, was used as a scaffold to design biosensors for naltrexone and quinine [16]. |
| Reporter Genes | Generate a measurable output (e.g., fluorescence, luminescence) linked to biosensor activity. | GFP for fluorescence-based screening and quantification via flow cytometry or plate readers [23] [10]. |
| Synthetic Promoters | Engineered DNA sequences that control transcription initiation in response to a specific TF. | The synthetic promoter QT was designed to be controlled by the artificial TF ProB [23]. |
| Ribosome Binding Site (RBS) Libraries | A diverse collection of RBS sequences with varying strengths to tune translation initiation. | Fine-tuning the expression level of a pathway enzyme to balance metabolic flux and reduce burden. |
| Small RNAs (sRNAs) | Used for post-transcriptional regulation by targeting specific mRNAs for silencing. | Implementing a post-transcriptional controller to enhance the evolutionary longevity of a gene circuit [24]. |
| Puromycin | An aminoacyl-tRNA analog that incorporates into nascent chains and halts translation. | Used in the SUnSET assay to label and quantify global protein synthesis rates [22]. |
| Ribo-tag / TRAP System | Enables cell-type-specific isolation of translating mRNAs from complex systems or in vivo models. | Studying the translatome of cardiomyocytes within heart tissue to understand cardiac disease [22]. |
| Cbz-Gly-Pro-Ala-O-cinnamyl | Cbz-Gly-Pro-Ala-O-cinnamyl, MF:C27H31N3O6, MW:493.6 g/mol | Chemical Reagent |
| SORT-PGRN interaction inhibitor 3 | SORT-PGRN interaction inhibitor 3, MF:C15H19Cl2NO3, MW:332.2 g/mol | Chemical Reagent |
Transcription factor-based biosensors (TFBs) are genetically encoded devices that utilize allosteric transcription factors (aTFs) to sense intracellular metabolite concentrations and convert this biological signal into a measurable output, typically fluorescence [8] [10]. This fundamental operating principle has positioned them as powerful tools for high-throughput screening (HTS) in synthetic biology and metabolic engineering. They enable researchers to rapidly sift through vast libraries of engineered microbial strains to identify rare high-performers, overcoming a critical bottleneck in the development of efficient microbial cell factories [25] [26]. By linking the production of a desired compound, such as a natural product or bio-chemical, to a simple fluorescence readout, TFBs transform the complex task of quantifying metabolite titers into a straightforward process of measuring fluorescence intensity, either in liquid culture or directly from colonies on agar plates [27] [14]. This review details the core mechanisms, applications, and methodologies for implementing TFB-driven HTS in strain and enzyme engineering programs.
Core Principles of TF-Based Biosensors for HTS
Mechanism of Action
The operation of a TFB follows a coherent logic that can be broken down into three key steps, as illustrated in the diagram below.
- Analyte Recognition: An intracellular transcription factor (TF) specifically binds to a target metabolite (the inducer or ligand), causing a conformational change in the TF [8].
- Signal Transduction: This ligand-induced change alters the TF's affinity for its specific DNA operator sequence located within a synthetic promoter. Depending on the biosensor design (activator- or repressor-based), this binding either initiates or relieves repression of transcription [8] [10].
- Output Generation: The change in promoter activity regulates the expression of a downstream reporter gene, such as Green Fluorescent Protein (GFP) or Red Fluorescent Protein (RFP). The fluorescence intensity of the cell population becomes a quantifiable proxy for the intracellular concentration of the target metabolite [8] [27].
Key Performance Metrics for HTS
For a TFB to be effective in HTS, it must be optimized for specific performance characteristics. A suboptimal biosensor can lead to high rates of false positives or negatives, failing to identify the best producers.
Table 1: Key Performance Metrics for HTS-Optimized Biosensors
| Metric | Definition | Impact on HTS | Ideal Value/Characteristic |
|---|---|---|---|
| Dynamic Range | The fold-change in output signal between the fully induced and uninduced (basal) states [8]. | A large dynamic range improves the resolution between high- and low-producing strains, making them easier to separate. | >10-fold to 100-fold is often desirable. |
| Sensitivity | The lowest concentration of the target metabolite that produces a detectable signal change, often quantified as the limit of detection (LOD) or the AC50 (concentration for half-maximal response) [25] [28]. | Determines the threshold of production that can be detected. Must be matched to the expected production levels of the library. | Low LOD and appropriate AC50 for the application. |
| Specificity | The ability of the biosensor to respond exclusively to the target metabolite and not to structurally similar analogs or other cellular components [8] [14]. | High specificity ensures that screening is based on the product of interest and not a side product, leading to more accurate selection. | Minimal cross-reactivity with other compounds. |
| Robustness | The consistency of biosensor performance across different genetic backgrounds, growth phases, and environmental conditions [26]. | Reduces context-dependent variability, ensuring that fluorescence reliably correlates with production titer across the entire library. | Stable performance under screening conditions. |
Application in HTS for Metabolic Engineering
TFBs are primarily deployed in two HTS paradigms: FACS-based screening for ultra-high-throughput analysis of millions of variants, and microtiter plate-based screening for smaller libraries or when using non-fluorescent reporters.
Case Studies in Bioproduction
The power of TFB-driven HTS is best illustrated by recent successful applications in engineering strains for valuable chemicals.
Table 2: Representative Case Studies of TFB-Driven HTS
| Target Product | Biosensor Core | Engineering Challenge | HTS Outcome & Strain Performance |
|---|---|---|---|
| Caffeic Acid (CA) [25] | Engineered CarR TF (from Acetobacterium woodii), responsive to p-coumaric acid (p-CA), a precursor to CA. | Low heterologous enzyme activity and product toxicity. | A 6.85-fold enhancement in catalytic activity of a key enzyme (FjTAL) was achieved. The final engineered strain (CA8) produced 9.61 g/L of CA in a bioreactor, the highest reported titer. |
| 5-Aminolevulinic Acid (5-ALA) [27] | Artificially evolved AsnC TF (from E. coli), mutated to respond to 5-ALA instead of its native ligand, L-asparagine. | No naturally occurring TF for 5-ALA was known, preventing direct screening. | The developed biosensor (EAC103-3H) had a linear detection range of 1-12 mM and was used to screen mutant libraries, leading to a 4.78-fold enhancement in 5-ALA production. |
| General Natural Products [14] | Various native or engineered TFs. | Low titers and yields in heterologous hosts. | The review highlights that TFBs enable high-throughput screening, adaptive evolution, and dynamic control to overcome metabolic bottlenecks in natural product synthesis. |
Experimental Workflow for HTS
A typical workflow for employing a TFB in a strain engineering campaign is methodical and iterative. The diagram below outlines the key stages from library creation to strain validation.
- Library Construction: Genetic diversity is introduced through methods like error-prone PCR, site-saturation mutagenesis of key pathway enzymes, or combinatorial assembly of pathway gene variants [25] [27].
- Biosensor Integration: The engineered TFB, encoded on a plasmid or integrated into the chromosome, is transformed into the mutant library. The biosensor's reporter gene (e.g., GFP) is transcriptionally linked to the production of the target metabolite.
- Cultivation & Expression: The library is cultivated in a format suitable for screening, such as 96- or 384-well microtiter plates or on solid agar plates. The culture conditions are optimized to allow for metabolite production and biosensor response.
- High-Throughput Sorting:
- Fluorescence-Activated Cell Sorting (FACS): For liquid cultures, this is the preferred method for analyzing millions of cells. Cells are passed through a flow cytometer, and individual cells with fluorescence intensities above a pre-set threshold (indicating high metabolite production) are physically sorted into collection tubes [25].
- Microtiter Plate-Based Screening: The fluorescence of each well in a microtiter plate is read with a plate reader. The top-performing wells (e.g., the top 1%) are selected for further analysis.
- Colony-Based Screening: For libraries grown on agar plates, colonies exhibiting the highest fluorescence (or most intense color) under appropriate imaging are manually picked.
- Validation & Scale-Up: The isolated candidate strains are re-cultured, and their production capability is rigorously validated using gold-standard analytical methods like HPLC or LC-MS/MS to confirm that the fluorescence signal accurately correlates with high product titer [25] [27]. The best confirmed hits are then used for further rounds of engineering or scaled-up production in bioreactors.
Computational and Protein Engineering for Advanced Biosensor Design
A significant challenge in applying TFBs is the limited availability of native TFs for many valuable, non-native metabolites. Two primary strategies are employed to overcome this limitation:
- Homology-Based Prediction & AI: Computational tools mine genomic and metagenomic databases to identify new TFs based on sequence homology to known regulator families (e.g., TetR, LysR, AraC) [10]. Artificial intelligence (AI) tools like DeepTFactor can predict novel TFs from genomic sequences, expanding the repertoire of potential biosensor components [10] [14].
- Directed Evolution of TFs: When a native TF for the desired compound does not exist, a TF with known structure and specificity can be engineered. This involves:
- Library Creation: Generating a library of TF mutants via site-saturation mutagenesis of key amino acid residues in the ligand-binding pocket [27].
- Positive-Negative Screening: Screening the mutant library for clones that have gained responsiveness to the new target ligand (positive selection) while having lost responsiveness to the native ligand (negative selection) [10] [27]. The case of the engineered AsnC TF for 5-ALA is a prime example of this approach [27].
The Scientist's Toolkit: Essential Research Reagents and Solutions
The development and implementation of a TFB-HTS platform require a suite of specialized reagents and tools.
Table 3: Essential Reagents and Materials for TFB-HTS
| Category / Reagent | Specific Examples | Function / Application |
|---|---|---|
| Chassis Organisms | Escherichia coli (DH5α, BL21), Bacillus subtilis, Saccharomyces cerevisiae | Model host organisms for heterologous pathway expression and biosensor operation [25] [27]. |
| Reporter Genes | GFP (Green Fluorescent Protein), RFP (Red Fluorescent Protein), other fluorescent proteins, Luciferase | Generates a quantifiable output signal linked to metabolite concentration [8] [27]. |
| Cloning & Expression | Plasmid vectors, Restriction enzymes, DNA ligases, Gibson Assembly mix, PCR reagents | For the construction of biosensor genetic circuits and metabolic pathways. |
| Screening Equipment | Flow Cytometer / FACS, Microtiter Plate Reader, Automated Colony Picker | Essential instrumentation for high-throughput measurement and sorting based on the biosensor signal [25]. |
| Analytical Validation | HPLC, LC-MS/MS, Ehrlich's reagent (for 5-ALA) | Gold-standard methods for validating the titer of the target product in screened strains [27]. |
| Target Metabolites | p-Coumaric acid, 5-Aminolevulinic acid, various natural products | Pure chemical standards for biosensor characterization, calibration, and use as positive controls [25] [27]. |
| Iso-phytochelatin 2 (Glu) | Iso-phytochelatin 2 (Glu)|For Research | |
| Antileishmanial agent-27 | Antileishmanial agent-27, MF:C24H16BrNO3S2, MW:510.4 g/mol | Chemical Reagent |
Transcription factor-based biosensors represent a paradigm shift in high-throughput screening for metabolic engineering. By providing a direct, functional link between a strain's production phenotype and a simple fluorescent genotype, they dramatically accelerate the design-build-test-learn cycle. While challenges remain in broadening the ligand scope and ensuring robustness, the continued integration of protein engineering, directed evolution, and computational/AI-based design promises to further expand the utility of these powerful tools. As exemplified by the success in producing compounds like caffeic acid and 5-ALA, TFB-driven HTS is an indispensable strategy for advancing the microbial production of valuable chemicals.
Transcription factor-based biosensors (TFBs) are genetically encoded devices that sense intracellular metabolite concentrations and convert this biological signal into a measurable output, typically gene expression [9] [8]. This functionality makes them powerful tools for implementing dynamic control in microbial cell factories. Unlike static metabolic engineering strategies, which often rely on constitutive gene expression, dynamic regulation allows a cell to autonomously adjust its metabolic flux in real-time, responding to the actual physiological state during fermentation [29]. This is crucial for resolving the fundamental conflict between cell growth and product formation, optimizing the metabolic state for each fermentation stage, and preventing the toxic accumulation of intermediates, thereby enhancing the production of valuable natural products and chemicals [14] [30].
Within the broader research context of transcription factor-based biosensors for dynamic regulation, this application specifically focuses on their capacity to function as closed-loop feedback control systems. These systems sense key intracellular metabolites and, based on their concentration, dynamically regulate the expression of pathway genes to maintain metabolic balance and maximize yield and productivity [8] [29]. This guide provides a technical deep-dive into the core principles, engineering methodologies, and experimental protocols for deploying TFBs in dynamic metabolic regulation.
Core Principles and Mechanisms of Action
The operational mechanism of a TFB for dynamic regulation involves a series of sequential steps that translate metabolite concentration into a tuned metabolic output.
Figure 1: The core mechanism of a transcription factor-based biosensor for dynamic metabolic regulation. The process involves sensing (red), signal transduction (blue), and genetic regulation (green) to achieve a final metabolic outcome (yellow).
- Analyte Recognition: An intracellular metabolite (the ligand or inducer, e.g., pyruvate) binds to a specific allosteric transcription factor (aTF), inducing a conformational change [9] [8].
- Signal Transduction: This conformational change alters the DNA-binding affinity of the aTF. Depending on the system design, this can result in the activation or derepression of a target promoter [8]. For instance, in a repressor-based system common in prokaryotes, ligand binding causes the repressor to detach from its operator site, allowing transcription to proceed.
- Output Generation: The change in promoter activity regulates the expression of a downstream gene. In dynamic metabolic engineering, this output gene is typically a key enzyme in a biosynthetic pathway. Its expression is therefore directly linked to the concentration of the sensed metabolite, creating a feedback loop [29].
This mechanism allows the construction of genetic circuits that can autonomously balance metabolism. For example, a biosensor can be designed to upregulate a product synthesis pathway only when a key precursor metabolite accumulates to high levels, thereby avoiding unnecessary metabolic burden during the growth phase [30] [29].
Engineering and Optimizing Biosensors for Dynamic Control
Native TFBs often lack the performance characteristics required for effective dynamic control. Thus, engineering is essential to optimize key properties as shown in the table below.
Table 1: Key performance properties of TF-based biosensors for dynamic regulation and strategies for their optimization.
| Property | Definition | Impact on Dynamic Regulation | Optimization Strategies |
|---|---|---|---|
| Sensitivity | The minimal ligand concentration that elicits a response [29]. | Determines the threshold for triggering regulation. | Site-directed mutagenesis of the ligand-binding pocket; screening natural homologs [29]. |
| Dynamic Range | The fold-change between the "ON" and "OFF" output states [9]. | Determines the strength of the regulatory response. | Engineering promoter sequences; tuning ribosomal binding sites (RBS); protein engineering of the TF [9] [8]. |
| Specificity | The ability to distinguish the target ligand from similar molecules [8]. | Prevents misregulation by off-target metabolites. | Structure-guided engineering of the ligand-binding domain [31]. |
| Leakiness | The baseline level of output gene expression in the absence of the ligand [29]. | Wastes cellular resources and can cause metabolic imbalance before induction. | Fine-tuning the affinity between the TF and its operator DNA sequence [29]. |
A prominent example is the engineering of a pyruvate-responsive biosensor based on the E. coli transcription factor PdhR. To optimize this system for controlling central metabolism, researchers screened multiple PdhR homologs from different bacteria to identify variants with superior intrinsic properties. This was followed by computational analysis and site-directed mutagenesis to further refine the ligand-binding pocket, resulting in a biosensor with reduced leakage, enhanced sensitivity, and a wider dynamic range [29]. Such an engineered biosensor was successfully applied to dynamically regulate the biosynthesis of trehalose and 4-hydroxycoumarin, leading to yield improvements of 2.33-fold and 1.63-fold, respectively, compared to static controls [29].
For metabolites without a known natural TF, de novo design of artificial transcription factors is an emerging approach. As demonstrated for the steroid androst-4-ene-3,17-dione (AD), this involves using molecular dynamics (MD) simulations to model a ligand-binding domain (LBD) and guide the assembly of a functional TF from modular parts: a DNA-binding domain (e.g., LexA), a flexible linker, and a transcriptional activation domain (e.g., B42) [31].
Experimental Workflow for Implementing Dynamic Regulation
Implementing a dynamic control strategy is a multi-stage process, from initial sensor characterization to final bioproduction validation. The following workflow and diagram outline the key stages.
Figure 2: A comprehensive experimental workflow for developing and implementing a dynamic metabolic regulation system, from initial sensor characterization to final validation.
Protocol: Biosensor Characterization and Application
Objective: To characterize the dose-response of a metabolite-responsive TFB and apply it for dynamic regulation of a target pathway.
Materials:
- Strains: An appropriate microbial host (e.g., E. coli BW25113 or other production chassis).
- Plasmids: A biosensor plasmid containing the TFB with a reporter gene (e.g., GFP), and a separate production plasmid for the target pathway.
- Chemicals: The target ligand (inducer), culture media (e.g., LB, M9 minimal medium), and appropriate antibiotics.
Methods:
Dose-Response Characterization:
- Transform the biosensor plasmid into the host strain.
- Inoculate cultures in media with varying concentrations of the target ligand.
- Measure the output signal (e.g., fluorescence) and cell density (OD₆₀₀) at regular intervals during growth.
- Plot the normalized output (e.g., fluorescence/OD₆₀₀) against the ligand concentration to generate a dose-response curve. From this curve, calculate the dynamic range (maximum output/minimum output) and the EC₅₀ (ligand concentration at half-maximal response) [29].
Genetic Circuit Assembly for Dynamic Control:
- Replace the reporter gene in the biosensor construct with a key enzyme gene from the target biosynthetic pathway. This creates a genetic circuit where the pathway gene is directly controlled by the metabolite sensor.
- Alternatively, the circuit can be designed to repress a competing pathway, thereby redirecting flux toward the desired product [29].
Bioproduction Evaluation:
- Engineer the production host by introducing the dynamic control circuit and the necessary production pathway genes.
- Perform fermentations in shake flasks or bioreactors. For central metabolism precursors like pyruvate, a fed-batch process with controlled carbon source feeding is often beneficial [29].
- Quantify the final product titer using analytical methods like HPLC or GC-MS and compare it against control strains using static regulation (e.g., constitutive expression).
Protocol: Quantitative Metabolomics for Validation
Objective: To validate the metabolic effects of dynamic regulation by quantifying changes in intracellular metabolite concentrations.
Materials:
- Stable Isotope-Labeled Internal Standard Mixture (SILIS): Prepared from cells grown on a fully ¹³C-labeled carbon source (e.g., U–¹³C₆-glucose) [30].
- Quenching Solution: Cold methanol.
- Extraction Solvent: Methanol/water or acetonitrile/methanol/water mixtures.
Methods:
Sample Collection and Metabolite Extraction:
- Rapidly quench culture samples in cold methanol (-40°C) to instantly halt metabolism.
- Centrifuge to pellet cells.
- Extract intracellular metabolites using an appropriate solvent system.
- Mix the extracted sample with a known amount of SILIS [30].
LC-MS/MS Analysis and Data Processing:
- Analyze the samples using Liquid Chromatography coupled with Tandem Mass Spectrometry (LC-MS/MS).
- Use the SILIS for the Stable Isotope Dilution Method (SIDM) to achieve absolute quantification of metabolite pools. The ¹³C-labeled internal standards correct for variations in extraction efficiency and ionization suppression during MS analysis [30].
- Compare the metabolite profiles (e.g., of the central precursor and pathway intermediates) between the dynamically regulated strain and control strains to confirm the intended redirection of metabolic flux.
Application Case Studies in Bioproduction
The following table summarizes successful applications of dynamic metabolic regulation for the production of various compounds, highlighting the key metabolites sensed and the outcomes achieved.
Table 2: Representative case studies of dynamic metabolic regulation in bioproduction.
| Product | Sensed Metabolite | Transcription Factor | Key Result | Reference |
|---|---|---|---|---|
| Trehalose | Pyruvate | PdhR (engineered) | 2.33-fold increase in titer (3.72 g/L) vs. static control. | [29] |
| 4-Hydroxycoumarin (4HC) | Pyruvate | PdhR (engineered) | 1.63-fold increase in titer (491.5 mg/L) vs. static control. | [29] |
| γ-Aminobutyric Acid (GABA) | Pathway Intermediates | N/A (Dynamic strategy applied) | Identification of metabolic bottlenecks via quantitative metabolomics. | [30] |
| Steroids (Androst-4-ene-3,17-dione) | Androst-4-ene-3,17-dione | AdT (Designed de novo) | Creation of a non-natural TF for a metabolite lacking a native sensor. | [31] |
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key research reagents, tools, and databases essential for developing TF-based biosensors for dynamic regulation.
| Category | Item | Function/Description | Example Sources/Tools |
|---|---|---|---|
| Biological Parts | Transcription Factor (TF) Genes | Sensing element for the target metabolite. | RegulonDB (for E. coli), P2TF, Pdb (for structures) [9]. |
| Reporter Genes | Provides a measurable output for sensor characterization. | GFP, RFP, Luciferase. | |
| Engineered Host Strains | Chassis for circuit implementation and production. | E. coli BW25113, XL1-Blue [29]. | |
| Computational Tools | Molecular Dynamics (MD) Simulation | Models TF-ligand interactions to guide design. | GROMACS, AMBER [31]. |
| Homology Modeling | Predicts protein structure based on known homologs. | SWISS-MODEL, ROSETTA3 [29]. | |
| Circuit Design Tools | Aids the in silico design of complex genetic circuits. | Cello [8]. | |
| Analytical Methods | LC-MS/MS with SILIS | Enables absolute quantification of intracellular metabolites. | [30] |
| Flow Cytometry | High-throughput characterization of biosensor response at single-cell level. | - | |
| Databases | TF-Ligand Databases | Catalog known interactions between TFs and small molecules. | RegulonDB, RegPrecise, PRODORIC [9]. |
| Bcat-IN-4 | BCAT-IN-4|BCAT Inhibitor|For Research | BCAT-IN-4 is a potent branched-chain aminotransferase (BCAT) inhibitor for cancer metabolism research. For Research Use Only. Not for human or diagnostic use. | Bench Chemicals |
| Zevotrelvir | Zevotrelvir, CAS:2773516-53-1, MF:C28H26F3N5O3, MW:537.5 g/mol | Chemical Reagent | Bench Chemicals |
Transcription factor-based biosensors (TFBs) are engineered genetic circuits that utilize natural sensing mechanisms to detect specific molecules and produce a measurable output. Their modular architecture, composed of a sensing element (the transcription factor) and an actuator module (a reporter gene), enables their customization for diverse applications [3]. This technical guide explores the cutting-edge deployment of these biosensors in clinical diagnostics and environmental monitoring, detailing the underlying mechanisms, experimental protocols, and key reagents that power this innovative technology.
Core Mechanisms and Signaling Pathways
At their core, TFBs function through a direct mechanism of analyte recognition and signal transduction. The pathway can be broken down into a series of sequential steps, as illustrated below.
The specific behavior of this pathway depends on whether the biosensor is designed around a transcriptional activator or a repressor:
- Activator-Based Systems: In the absence of the analyte, the activator TF does not bind DNA, and reporter gene expression is low. Upon analyte binding, the TF undergoes a conformational change that allows it to bind its operator site, recruiting RNA polymerase and activating transcription of the reporter gene [10] [32].
- Repressor-Based Systems: In the absence of the analyte, the repressor TF binds its operator site, physically blocking RNA polymerase and repressing transcription. Analyte binding induces a conformational change that causes the TF to dissociate from the DNA, thereby derepressing transcription and allowing reporter gene expression [10] [33].
Applications in Clinical Diagnostics
In clinical diagnostics, TFBs offer a path toward rapid, specific, and low-cost detection of pathogens and biomarkers. A primary application is the detection of specific microbial infections through quorum-sensing molecules. For instance, biosensors utilizing LuxR, a TF that recognizes acyl-homoserine lactones (AHLs) produced by many Gram-negative bacteria, have been developed as critical tools for identifying microbial infections [8]. Furthermore, biosensors for antibiotics like tetracycline (using TetR) and macrolides (using MphR) can be used both for monitoring therapeutic drug levels in patients and for identifying antibiotic-resistant pathogens [34] [8].
Table 1: Transcription Factor-Based Biosensors in Clinical Diagnostics
| Transcription Factor | Target Analyte | Host Chassis | Reported Output | Potential Diagnostic Application |
|---|---|---|---|---|
| LuxR [8] | Acyl-Homoserine Lactones (AHLs) | E. coli | Luminescence/GFP | Detection of Gram-negative bacterial infections |
| TetR [34] | Tetracycline | E. coli | Fluorescence | Monitoring drug levels; detecting antibiotic resistance |
| MphR [34] | Erythromycin, Azithromycin | E. coli | Fluorescence | Monitoring macrolide antibiotic concentrations |
| SoxR [35] | NADPH | E. coli | eYFP | Reporter for cellular metabolic state |
Experimental Protocol: Diagnostic Biosensor Workflow
The following workflow outlines the key steps for employing a TFB for diagnostic purposes, from initial culturing to signal measurement.
Detailed Methodology:
- Strain Preparation: Inoculate a liquid culture of the engineered biosensor strain (e.g., E. coli harboring the plasmid-borne LuxR-based biosensor circuit). Grow the culture to the mid-exponential phase (OD600 ~0.5-0.6) under appropriate antibiotic selection [35] [13].
- Sample Introduction: Aliquot the cell culture into a multi-well plate. Add the clinical sample (e.g., sputum, urine, or serum) or a standard solution of the target analyte (e.g., AHL for LuxR sensor) to the test wells. Use a negative control (e.g., culture medium) and positive controls (known analyte concentrations) on the same plate [13].
- Incubation: Incubate the plate under optimal growth conditions (e.g., 37°C for E. coli) for a defined period, typically 2-8 hours, to allow the analyte to enter the cells, interact with the TF, and induce reporter gene expression [13].
- Signal Measurement: Quantify the output signal using appropriate instrumentation.
- Data Analysis: Calculate the fold-change in output signal (test/control) and generate a dose-response curve by plotting the output signal against the analyte concentration of the standards. The analyte concentration in the unknown sample can be interpolated from this standard curve [13] [3].
Applications in Environmental Monitoring
TFBs are exceptionally suited for on-site, real-time monitoring of environmental pollutants. They have been successfully engineered to detect a wide range of contaminants, from heavy metals to industrial plastic monomers.
Table 2: Transcription Factor-Based Biosensors in Environmental Monitoring
| Transcription Factor | Target Analyte | Host Chassis | Reported Output | Environmental Application |
|---|---|---|---|---|
| MerR [8] | Mercury (Hg²âº) | E. coli | Luminescence/GFP | Detection of mercury contamination in water |
| ArsR [8] | Arsenic (As³âº/âµâº) | E. coli | Luminescence | Monitoring arsenic in groundwater |
| TphR [32] | Terephthalate (TPA) | E. coli | Fluorescence | Monitoring PET plastic degradation |
| XylR [2] | Xylose | E. coli | Fluorescence | Reporting on lignocellulosic biomass hydrolysis |
| BenR [35] | Benzoate | E. coli | GFP | Detection of aromatic hydrocarbon pollutants |
A notable advancement is the development of biosensors for the plastic monomer terephthalate (TPA). The TphR protein, an activator TF, has been mined bioinformatically and engineered into biosensors in E. coli. These biosensors are crucial for screening enzymes that degrade polyethylene terephthalate (PET) plastic and for monitoring environmental TPA levels [32]. Similarly, heavy metal sensors like MerR and ArsR have been deployed to detect mercury and arsenic in water samples with high sensitivity, achieving detection limits as low as 0.2 μg/L for mercury, demonstrating their field applicability for environmental risk assessment [8].
Experimental Protocol: Environmental Biosensor Workflow
The process for environmental monitoring often involves handling complex samples and may require pre-processing to make the analyte bioavailable to the sensor strain.
Detailed Methodology:
- Sample Collection & Preparation: Collect environmental samples (water, soil extract). For solid samples like soil, perform extraction using a suitable buffer. Filter water samples if necessary to remove large particulates and minimize microbial contamination. Adjust the sample pH to a range compatible with the biosensor strain (e.g., pH 7.0) [13].
- Biosensor Exposure: Combine the processed environmental sample with the biosensor cell culture in a ratio that maintains cell viability while maximizing analyte exposure. For liquid samples, this can be a direct addition to the culture medium [13] [32].
- Induction & Growth: Incubate the mixture with shaking at the optimal temperature for the host chassis. The duration must be optimized to allow for sufficient analyte uptake and reporter protein expression, which can range from a few hours to an entire growth cycle [32].
- High-Throughput Assay: For efficient screening of multiple samples or conditions, transfer the cultures to a multi-well plate. This format is compatible with automated liquid handlers and plate readers, significantly increasing throughput [32].
- Signal Quantification: Measure the output signal (fluorescence, luminescence) using a plate reader. Compare the signals from environmental samples to a standard curve generated with known concentrations of the target pollutant to determine the concentration in the sample [13] [32].
Performance Optimization and Tuning
The practical utility of a TFB is dictated by its performance characteristics. These parameters must be carefully tuned for the specific application, whether it requires detecting trace contaminants or operating over a wide concentration range.
Table 3: Key Performance Parameters for Transcription Factor-Based Biosensors
| Performance Parameter | Definition | Influencing Factors | Tuning Strategies |
|---|---|---|---|
| Sensitivity (Kâ‚/â‚‚) [34] [3] | The analyte concentration required for half-maximal response. | TF's intrinsic affinity for the ligand. | Directed evolution of the Ligand Binding Domain (LBD) [2]; RBS engineering to modulate TF expression levels [2]. |
| Dynamic Range [34] [3] | The ratio between the maximal (ON) and minimal (OFF) output signal. | Strength of TF-DNA and TF-ligand interactions. | Engineering the core promoter and operator sequence [32]; using stronger transcriptional activators [33]. |
| Operational Range [36] [3] | The concentration window over which the biosensor shows a graded response. | Dynamic range and cooperativity of the response. | Directed evolution to create TF variants with altered effector profiles [36]. |
| Specificity [2] | The ability to distinguish the target analyte from structurally similar molecules. | Structure and amino acid composition of the TF's ligand-binding pocket. | Structure-guided mutagenesis and high-throughput screening (e.g., using FACS) [34] [2]. |
| Response Time [3] | The time taken to reach a measurable output after analyte exposure. | Rates of transcription and translation of the reporter. | Using destabilized reporter proteins for faster turnover [3]. |
The Scientist's Toolkit: Research Reagent Solutions
The development and application of TFBs rely on a standardized set of biological tools and reagents.
Table 4: Essential Research Reagents for TFB Development
| Reagent / Tool | Function | Example(s) |
|---|---|---|
| Plasmid Vectors | Harbors the genetic circuit for the biosensor, including the TF gene and reporter gene. | High-copy (e.g., pUC origin) or low-copy (e.g., p15A origin) plasmids for modulating gene dosage [2]. |
| Reporter Genes | Encodes a measurable output protein that is expressed under the control of the TF. | Fluorescent Proteins (GFP, RFP, eYFP) [35] [33]; Luminescent Proteins (Luciferase); Antibiotic Resistance Genes (TetA) [35]. |
| Host Chassis | The microbial cell that houses the biosensor circuit and provides the necessary machinery for gene expression. | Escherichia coli (common, well-characterized) [35]; Pseudomonas putida (robust, for environmental apps) [13]; Saccharomyces cerevisiae (eukaryotic chassis) [33]. |
| TF Engineering Tools | Methods to alter the specificity and sensitivity of the transcription factor. | Site-directed mutagenesis kits; Error-prone PCR kits for random mutagenesis [2]. |
| High-Throughput Screening | Technology for rapidly sorting and isolating high-performing biosensor variants from large libraries. | Fluorescence-Activated Cell Sorting (FACS) [34] [2]. |
| Computational Design Software | In silico tools for predicting protein structure, designing mutations, and modeling circuit performance. | AlphaFold for TF structure prediction [10]; Cello for genetic circuit design [8]. |
| Ido1-IN-22 | IDO1-IN-22|Potent IDO1 Inhibitor|For Research Use | IDO1-IN-22 is a potent IDO1 inhibitor (IC50=17.6 nM). It blocks immunosuppressive kynurenine pathway in cancer research. For Research Use Only. Not for human consumption. |
| Egfr-IN-94 | Egfr-IN-94, MF:C19H17ClN8OS, MW:440.9 g/mol | Chemical Reagent |
Overcoming Practical Hurdles: A Guide to Biosensor Optimization and Problem-Solving
Transcription factor (TF)-based biosensors are indispensable tools in synthetic biology, enabling dynamic regulation of metabolic pathways and high-throughput screening of production strains. These genetic circuits, typically composed of a transcription factor and its cognate promoter regulating a reporter gene, allow cells to detect specific metabolites and elicit a programmed response [9]. However, their transition from proof-of-concept demonstrations to robust, reliable applications is often hindered by three persistent challenges: leaky expression, where low-level gene expression occurs in the absence of the inducer; cross-talk, where unintended interactions disrupt the specific signal; and host context effects, where the heterologous biosensor fails to function as expected in a new organism [9] [10]. This technical guide delves into the molecular origins of these challenges and provides a comprehensive framework of experimental and computational strategies to overcome them, thereby enhancing the performance and applicability of biosensors in dynamic regulation research.
Leaky Expression: Unwanted Background Noise
Leaky expression fundamentally stems from imperfect repression or activation in the "off" state. For repressor-based biosensors, this occurs when the transcription factor does not fully block RNA polymerase from initiating transcription at the target promoter. This can be due to a low binding affinity of the TF for its operator sequence, an insufficient intracellular concentration of the repressor itself, or competition with host-encoded factors that promote transcription [37] [9]. In activator-based systems, background noise can arise from the activator protein weakly stimulating transcription even in the absence of its ligand.
Cross-Talk: Loss of Signal Specificity
Cross-talk refers to the interference between the biosensor circuit and the host's native regulatory networks. This can manifest in several ways: the host's transcription machinery may not properly recognize the biosensor's regulatory elements, endogenous metabolites or TFs may inadvertently bind to and trigger the biosensor, or the biosensor may sequester essential host factors [9]. This lack of orthogonality compromises the specificity of the biosensor, leading to false positives or inaccurate readings of metabolite concentrations.
Host Context: The Challenge of Portability
A biosensor that functions perfectly in its native organism often performs poorly when transferred to a standard chassis like E. coli or S. cerevisiae. This host context problem includes issues such as improper protein folding, incompatible codon usage, differences in promoter recognition by the host's RNA polymerase, and unexpected interactions with the host's proteostasis network [37] [10]. Furthermore, the "housekeeping" context of a new host—its growth rate, metabolic burden, and intracellular environment—can significantly alter biosensor dynamics, including its operational and dynamic range [37].
Engineering Solutions for Robust Biosensor Performance
Strategic Genetic Refactoring
A powerful approach to mitigate these challenges is the systematic refactoring of the biosensor's genetic components.
Promoter and Operator Engineering: The performance of a biosensor is highly dependent on the interplay between the transcription factor and its cognate promoter. A unified design strategy involves replacing the native promoter controlling the TF gene with well-characterized, synthetic constitutive promoters that are tailored for the host organism [37]. This allows for fine-tuning of TF expression levels, which can directly impact leakiness and dynamic range. For instance, modifying the operator sequence within the inducible promoter can also enhance binding affinity and reduce leakiness. Tools like Snowprint can predict optimal operator sequences for previously uncharacterized TFs, facilitating this process [38].
Table 1: Genetic Components for Biosensor Refactoring
| Genetic Component | Engineering Strategy | Impact on Performance | Example |
|---|---|---|---|
| TF Expression Promoter | Replace native promoter with synthetic constitutive promoters of varying strength [37]. | Fine-tunes TF concentration; reduces leakiness, restores response in heterologous hosts [37]. | Fine-tuning TtgR expression to optimize flavonoid sensor response [7]. |
| Inducible Promoter | Modify operator sequence spacing, copy number, and sequence [32]. | Alters TF binding affinity; increases dynamic range, reduces background [32]. | Using Snowprint to predict high-affinity operators for TetR-family regulators [38]. |
| Ribosome Binding Site (RBS) | Engineer RBS strength to control translation efficiency [9]. | Optimizes TF translation; balances cellular resources, mitigates burden. | Not explicitly detailed in results, but standard practice in synthetic biology [9]. |
| Reporter Gene | Use rapid-maturation fluorescent proteins or enzymatic reporters (e.g., lacZ) [39]. | Improves signal-to-noise ratio; enables quantitative and visual readouts. | Using eGFP for fluorescence or lacZ for colorimetric output in biosensors [7] [39]. |
Protein Engineering of the Transcription Factor: When the native TF lacks the desired specificity or sensitivity, its ligand-binding pocket can be directly mutated. Structure-guided mutagenesis, informed by computational docking studies, can yield TF variants with altered sensing profiles.
- Example: The TtgR repressor from Pseudomonas putida was engineered to enhance its selectivity for specific flavonoids. The wild-type TtgR responds to a broad range of ligands. However, by introducing a single point mutation (N110F) into the ligand-binding pocket, researchers created a biosensor variant with a tailored response, enabling accurate quantification of resveratrol and quercetin [7].
Computational and Modeling Approaches
Advanced computational methods are becoming indispensable for the de novo design and optimization of biosensors.
- Predictive Tools: Snowprint is a bioinformatic tool that predicts the operator sequences for a given transcriptional regulator. By inputting a regulator's protein accession ID, Snowprint identifies conserved inverted repeat sequences in the regulator's genomic context, significantly accelerating the domestication of new biosensors [38].
- Design of Experiments (DoE): Instead of testing one variable at a time, a DoE approach allows for the efficient exploration of complex sequence-function relationships. This was successfully applied to engineer a TphR-based biosensor for terephthalic acid, where the core promoter and operator regions were systematically varied and modeled to produce biosensors with customized dynamic ranges, sensitivities, and steepness of response [32].
The following diagram illustrates a cohesive workflow that integrates these computational and experimental strategies to overcome core biosensor challenges.
The Scientist's Toolkit: Essential Reagents and Protocols
This section details key reagents and methodologies employed in the development and validation of engineered biosensors, as referenced in the studies.
Table 2: Research Reagent Solutions for Biosensor Engineering
| Reagent / Material | Function in Biosensor Development | Specific Example from Literature |
|---|---|---|
| TtgR Transcription Factor | Transcriptional repressor used as sensing element for flavonoids and antibiotics [7]. | Wild-type and mutant TtgR (e.g., N110F) used in E. coli-based biosensors for resveratrol and quercetin [7]. |
| FapR/FapR-AD System | Malonyl-CoA sensing repressor (FapR) or activator (FapR-AD) for dynamic pathway regulation [40]. | Used in yeast for dynamic control of fatty acid and 3-HP biosynthesis pathways [40]. |
| LasI-LasR Quorum Sensing Circuit | Sensing module for AHLs, applied in environmental detection [39]. | Engineered in E. coli with lacZ reporter for microplastic detection via AHL signaling [39]. |
| Constitutive Promoter Library | To control the expression level of the transcription factor gene [37]. | Fine-tuning biosensor response by selecting promoters of different strengths for TF expression [37]. |
| Reporter Genes (eGFP, lacZ) | Provides a measurable output (fluorescence, colorimetry) for biosensor activity [7] [39]. | eGFP for fluorescence quantification in TtgR biosensors [7]; lacZ for blue/white screening in LasR-based sensors [39]. |
| Alginate Hydrogel | Material for physical immobilization of whole-cell biosensors, enhancing stability [39]. | Used to encapsulate E. coli biosensor cells for colorimetric AHL detection [39]. |
Detailed Experimental Protocol: Characterizing a Novel Biosensor
The following workflow, adapted from multiple studies, outlines the key steps for constructing and characterizing a newly designed TF-based biosensor in a heterologous host [7] [37] [39].
Biosensor Construction:
- Molecular Cloning: Clone the gene for the transcription factor (wild-type or mutant) under a constitutive promoter into a plasmid. In a second plasmid (or a different location on the same plasmid), clone the cognate inducible promoter, driving the expression of a reporter gene (e.g., eGFP). The inducible promoter should contain the predicted or known operator sequence. Restriction enzymes or Gibson assembly are commonly used [7].
- Transformation: Co-transform both plasmids into the heterologous host, typically E. coli BL21(DE3) or a similar laboratory strain [7].
Functional Assay & Characterization:
- Culture Preparation: Inoculate main cultures of the biosensor strain in a defined medium in a multi-well plate (e.g., a 48-well FlowerPlate) [37].
- Induction: Expose the biosensor cells to a range of concentrations of the target ligand. Include controls with no ligand (to measure leakiness) and with potential interfering compounds (to assess cross-talk) [7] [39].
- Output Measurement: After a defined incubation period, measure the output signal. For fluorescent reporters like eGFP, use a plate reader or flow cytometry. For enzymatic reporters like lacZ, add a substrate (e.g., X-Gal) and quantify the color development over time using a plate reader or image analysis software like ImageJ [7] [39].
Data Analysis: Calculate the biosensor's key performance parameters:
- Dynamic Range: The ratio (or fold-change) between the output signal at a saturating ligand concentration and the output signal in the absence of ligand.
- Leakiness: The output signal in the absence of ligand.
- Sensitivity/EC50: The concentration of ligand that produces a half-maximal response.
- Specificity: Assessed by the biosensor's response to non-target ligands that are structurally similar or relevant to the host's metabolism [7] [37].
The strategic engineering approaches outlined in this guide—from genetic refactoring and protein design to computational prediction—provide a robust pathway to overcome the classic challenges of leakiness, cross-talk, and host context in TF-based biosensors. The field is moving toward a more predictive and automated design cycle, powered by tools like Snowprint and DoE, which will dramatically accelerate the development of reliable, high-performance biosensors. As these tools mature, the application of robust, dynamically regulated biosensors will become routine, pushing the boundaries of metabolic engineering, advanced diagnostics, and the development of sophisticated engineered living materials.
Transcription factor (TF)-based biosensors are powerful tools in synthetic biology and metabolic engineering, capable of converting the intracellular concentration of specific small molecules into measurable genetic outputs [10] [8]. These biological devices typically consist of a sensing element—an allosteric transcription factor that undergoes conformational change upon ligand binding—and a reporting element that produces a detectable signal such as fluorescence, luminescence, or enzyme activity [10]. This signal transduction mechanism enables real-time monitoring of metabolic fluxes, high-throughput screening of production strains, and dynamic regulation of biosynthetic pathways [14] [8].
However, the native effector specificity of naturally occurring transcription factors often limits their immediate application in engineered systems [41] [10]. Many desirable target metabolites lack cognate sensing elements, while existing TFs may exhibit cross-reactivity with structurally similar compounds or insufficient sensitivity for the intended application [7]. Directed evolution has emerged as a powerful protein engineering strategy to overcome these limitations by artificially mimicking natural evolutionary processes in laboratory settings [41]. This approach involves iterative rounds of mutagenesis and screening to generate transcription factor variants with altered ligand specificity, enhanced sensitivity, or improved operational range [36].
Within the broader context of transcription factor-based biosensors for dynamic regulation research, engineering the sensing element represents a critical foundational step. By reprogramming TF specificity, researchers can create tailored biosensing systems that respond to non-native ligands, thereby expanding the toolbox available for metabolic engineering, synthetic biology, and therapeutic development [7] [14]. This technical guide examines the methodologies, applications, and recent advances in directed evolution of transcription factors, with particular emphasis on practical implementation for research and development.
Fundamental Principles of Transcription Factor-Based Biosensors
Architecture and Mechanism of Action
Transcription factor-based biosensors operate through a coordinated mechanism involving three fundamental steps: analyte recognition, signal transduction, and output generation [8]. In the initial recognition phase, the transcription factor binds to a specific target molecule (effector or ligand), inducing a conformational change that alters its DNA-binding affinity [10]. This molecular recognition event is highly specific, relying on complementary structural and chemical interactions between the TF's binding pocket and the ligand [7].
The signal transduction phase involves the TF's altered interaction with specific DNA operator sequences within promoter regions. Depending on the TF's native function—as either an activator or repressor—this interaction either facilitates or obstructs RNA polymerase binding, thereby modulating transcription of downstream reporter genes [10]. The final output generation phase produces a quantifiable signal corresponding to ligand concentration, typically through expression of fluorescent proteins, enzymes catalyzing colorimetric reactions, or selectable markers [8].
Table 1: Major Transcription Factor Families Used in Biosensor Development
| TF Family | Representative Effectors | Regulatory Mechanism | Example Applications |
|---|---|---|---|
| TetR | Antibiotics, Flavonoids | Repression | Metabolic engineering, High-throughput screening [10] |
| MerR | Heavy metals, Antibiotics | Activation | Environmental monitoring, Dynamic regulation [8] |
| AraC | Sugars, Aromatic compounds | Activation/Repression | Bioproduction optimization [10] |
| LysR | Amino acids, Intermediates | Activation | Pathway regulation, Metabolic flux control [10] |
| LuxR | Acyl-homoserine lactones | Activation | Quorum sensing, Microbial communication [8] |
Key Performance Parameters for Biosensor Engineering
Evaluating the performance of transcription factor-based biosensors requires assessment of several critical parameters that directly impact their utility in practical applications. Specificity refers to the biosensor's ability to distinguish the target ligand from structurally similar compounds, a property determined by the molecular complementarity between the TF's binding pocket and the ligand [7]. Sensitivity describes the minimum ligand concentration that elicits a detectable response, typically quantified as the limit of detection (LOD) or the effective concentration for half-maximal response (EC50) [36].
The dynamic range represents the ratio between the maximum and minimum output signals across the operational ligand concentration range, determining the biosensor's ability to discriminate between different metabolite levels [36]. Orthogonality ensures that the biosensor functions independently of host regulatory networks, minimizing interference with native cellular processes [10]. Finally, operational robustness refers to consistent performance across varying environmental conditions, including pH, temperature, and growth phase [8].
Directed Evolution Strategies for Transcription Factor Engineering
Library Creation Methods
Directed evolution begins with the generation of genetic diversity within the transcription factor gene, creating variant libraries that can be screened for desired properties. Several mutagenesis strategies offer distinct advantages depending on the available structural and functional information about the target TF.
Random mutagenesis employs error-prone PCR or mutator strains to introduce mutations throughout the entire TF coding sequence, requiring no prior knowledge of the protein's structure-function relationships [42]. This approach explores a vast sequence space but typically necessitates high-throughput screening due to the low probability of beneficial mutations [42]. Site-saturation mutagenesis targets specific residues within the ligand-binding pocket for comprehensive amino acid substitution, focusing diversity on regions most likely to influence effector recognition [7]. This method significantly reduces library size while increasing the probability of identifying functional variants.
Homology-based engineering leverages evolutionary relationships between transcription factors from different organisms, using multiple sequence alignments to identify variable regions associated with functional divergence [10]. Structure-guided mutagenesis utilizes computational or experimental protein structures to rationally select residues for mutagenesis based on their spatial arrangement within the binding pocket [7]. Recent advances in machine learning-based structure prediction, particularly AlphaFold, have dramatically accelerated this approach by providing reliable protein models without experimental structure determination [10] [42].
Directed Evolution Workflow for Transcription Factor Engineering
Case Studies in TF Directed Evolution
Engineering MphR for Macrolactone Aglycone Detection
The macrolide-sensing transcription factor MphR naturally responds to glycosylated macrolides but exhibits minimal sensitivity to their aglycone precursors, limiting its application in polyketide synthase (PKS) engineering [41]. Researchers addressed this limitation through an "effector walking" strategy, gradually shifting MphR's specificity from the native effector (macrolides with desosamine sugar) toward macrolactone aglycones through successive rounds of mutagenesis and screening [41]. This approach was combined with efflux pump deletion to increase intracellular aglycone concentrations, enhancing screening efficiency. The resulting MphR variants demonstrated dramatically broadened effector profiles that included several erythronolide macrolactones, enabling their application in high-throughput screening of PKS variants [41].
Optimizing CaiF for l-Carnitine Detection
CaiF, a transcriptional activator of l-carnitine metabolism, was engineered to extend its dynamic and operational range [36]. Researchers employed computational structural analysis to model CaiF's DNA binding site, followed by alanine scanning to identify key functional residues [36]. A "Functional Diversity-Oriented Volume-Conservative Substitution Strategy" was then applied to these residues, generating variants with altered dynamic ranges. The optimized CaiF variant (CaiF-Y47W/R89A) exhibited a 1000-fold wider concentration response range (10â»â´ mM to 10 mM) and a 3.3-fold higher output signal intensity compared to the wild-type biosensor [36].
Reprogramming TtgR for Flavonoid Sensing
TtgR from Pseudomonas putida natively regulates multidrug resistance efflux pumps and exhibits broad ligand specificity [7]. To enhance its selectivity for specific flavonoids, researchers performed structure-guided engineering of the TtgR binding pocket, generating point mutants at residues predicted to interact with ligands [7]. Computational structural analysis and ligand docking simulations informed rational mutagenesis, resulting in TtgR variants with altered sensing profiles. Notably, the N110F mutant enabled specific detection of resveratrol and quercetin with greater than 90% accuracy at 0.01 mM concentration, demonstrating how targeted engineering can refine TF specificity [7].
Table 2: Quantitative Performance Metrics of Engineered Transcription Factors
| Transcription Factor | Engineering Strategy | Key Mutations | Performance Improvement |
|---|---|---|---|
| MphR [41] | Effector walking + efflux pump deletion | Not specified | Dramatically broadened effector profile to include macrolactone aglycones |
| CaiF [36] | Computational design + volume-conservative substitution | Y47W/R89A | 1000-fold wider response range (10â»â´ mM–10 mM), 3.3-fold higher signal |
| TtgR [7] | Structure-guided binding pocket engineering | N110F | >90% accuracy for resveratrol and quercetin detection at 0.01 mM |
| ZntR [8] | Chimeric MerR-family engineering | Not specified | Enhanced metal specificity for Pb detection |
Experimental Protocols for Directed Evolution
High-Throughput Screening Methodologies
Effective directed evolution requires robust screening methods capable of evaluating large variant libraries. Fluorescence-Activated Cell Sorting (FACS) enables quantitative, single-cell resolution screening by coupling TF activation to fluorescent protein expression [8]. This method offers exceptionally high throughput (up to 10⸠cells per day) and direct quantitative assessment of biosensor performance across a wide dynamic range [8].
Microfluidic droplet screening encapsulates individual library variants in water-in-oil emulsions along with reporter systems and effectors, enabling ultra-high-throughput compartmentalized assays [42]. This approach is particularly valuable for detecting gaseous or volatile hydrocarbons that would otherwise diffuse in open systems [42]. Colony-based fluorescence screening provides a more accessible alternative, where library variants are plated on solid media and screened using automated imaging systems to quantify fluorescence intensity in response to effector addition [7].
For transcription factors that activate endogenous stress response pathways, growth-coupled selection offers a powerful screening strategy by linking TF activation to essential gene expression [42]. This method enables direct selection of functional variants without specialized equipment, as cells containing active biosensors outcompete non-functional variants during cultivation [42].
Characterization of Engineered Transcription Factors
Comprehensive characterization of evolved TF variants is essential to validate directed evolution outcomes. Dose-response analysis determines key performance parameters including dynamic range, sensitivity (EC50), and background expression (leakiness) by measuring output signals across a range of effector concentrations [7] [36]. This analysis typically employs fluorescence measurements, luminescence assays, or colorimetric readouts in a plate reader format.
Specificity profiling evaluates cross-reactivity with structurally similar compounds to ensure the engineered TF meets application requirements [7]. For the engineered TtgR variants, this involved testing response against multiple flavonoids (naringenin, quercetin, phloretin) and resveratrol to demonstrate altered specificity profiles [7]. Orthogonality assessment verifies that the engineered biosensor operates independently of host regulatory networks by testing function across different genetic backgrounds and growth conditions [10].
Binding affinity measurements provide direct quantitative analysis of TF-effector interactions using techniques such as Surface Plasmon Resonance (SPR), Microscale Thermophoresis (MST), or Isothermal Titration Calorimetry (ITC) [43]. These biophysical methods yield precise dissociation constants (Kd) and thermodynamic parameters that complement cellular biosensor data [43].
Table 3: Key Research Reagent Solutions for TF Directed Evolution
| Reagent/Resource | Function | Example Application |
|---|---|---|
| Error-prone PCR kits | Introduce random mutations throughout TF coding sequence | Generating diverse variant libraries without structural information [42] |
| Site-directed mutagenesis kits | Create specific amino acid substitutions | Targeted engineering of binding pocket residues [7] |
| Fluorescent reporter plasmids | Quantify TF activation via measurable output | FACS screening of library variants [7] [8] |
| Microfluidic droplet generators | Compartmentalize single cells for ultra-high-throughput screening | Detection of gaseous or volatile hydrocarbon production [42] |
| Protein structure prediction tools (AlphaFold) | Generate reliable protein models from sequence data | Structure-guided mutagenesis without experimental structure [10] [42] |
| TF-specific databases (RegTransBase, ColiNet) | Access curated information on TF-DNA interactions | Identifying natural TF candidates for engineering [10] |
| Ligand docking software | Simulate TF-effector interactions in silico | Predicting mutation effects on binding affinity [7] |
Visualization of Key Signaling Pathways and Regulatory Mechanisms
TF Biosensor Mechanism in Repressor-Type Systems
Applications in Metabolic Engineering and Therapeutic Development
Engineered transcription factor-based biosensors have enabled significant advances across multiple biotechnology domains. In natural product biosynthesis, dynamically regulated biosensors optimize metabolic flux by coupling pathway intermediate concentrations to expression of bottleneck enzymes [14] [8]. This closed-loop control system automatically adjusts enzyme levels in response to metabolite accumulation, overcoming kinetic imbalances that limit product yields [14].
For high-throughput strain development, engineered biosensors enable rapid screening of mutant libraries by linking desired phenotypic traits (e.g., product titers) to easily measurable outputs [41] [8]. The evolved MphR variants, for instance, allow identification of productive polyketide synthase variants from combinatorial libraries by detecting intracellular aglycone concentrations [41]. This approach dramatically accelerates the design-build-test-learn cycle in metabolic engineering.
In therapeutic development, engineered biosensors serve as diagnostic tools that detect disease biomarkers or regulate therapeutic gene expression in response to pathological signals [8]. Biosensors based on LuxR have been engineered to detect microbial infections by sensing acyl-homoserine lactones, demonstrating potential for clinical diagnostics [8]. Additionally, biosensors responsive to disease-specific metabolites enable precise temporal and spatial control of therapeutic gene expression in advanced cell-based therapies.
Future Perspectives and Emerging Technologies
The integration of artificial intelligence and machine learning represents the next frontier in transcription factor engineering [10] [14]. AI-assisted approaches can predict mutation effects on TF specificity and sensitivity, dramatically reducing experimental screening requirements [14]. Tools like DeepTFactor predict transcription factors from genomic sequences, while RoseTTAFold and AlphaFold enable accurate structure prediction to guide rational design [10] [42].
Cell-free biosensing systems employing isolated TF-based components offer advantages for detecting toxic compounds or operating in complex environments incompatible with living cells [14]. These systems leverage the specificity of engineered TFs while eliminating constraints associated with cellular viability, membrane permeability, and efflux pumping [14].
Expanding biosensor applications to non-model microorganisms will unlock new possibilities for industrial biotechnology by exploiting the unique metabolic capabilities of unconventional hosts [14]. This requires developing genetic tools and engineered biosensors tailored to these organisms, enabling real-time monitoring and control of bioprocesses in industrially relevant hosts [14].
Finally, multi-input biosensor networks that integrate multiple engineered TFs will enable sophisticated computation and decision-making in synthetic biology applications [8]. These systems can process complex environmental signals and implement dynamic control strategies that optimize bioproduction processes or diagnostic accuracy through integrated signal processing [8].
Directed evolution of transcription factor-based biosensors continues to expand the possibilities for metabolic engineering, therapeutic development, and synthetic biology. By applying the principles and methodologies outlined in this technical guide, researchers can engineer sensing elements with customized properties, enabling precise monitoring and control of biological systems for diverse applications.
In the design of transcription factor (TF)-based biosensors for dynamic regulation, achieving precise control over genetic circuits is paramount. These biosensors, which typically consist of a sensing component (an allosteric transcription factor) and a reporter component, enable cells to detect specific chemical compounds and elicit a programmed response [10]. The performance of these sophisticated genetic devices is critically dependent on the fine-tuning of their underlying genetic components, particularly promoters and ribosome binding sites (RBSs). Engineered biosensors serve vital functions in high-throughput screening of production strains and implementing dynamic regulation strategies in metabolic engineering and therapeutic applications [10] [3]. The careful engineering of promoters and RBSs allows researchers to optimize key biosensor performance parameters, including dynamic range, operating range, response time, and signal-to-noise ratio [3], ultimately determining the reliability and applicability of these systems in real-world settings.
Core Promoter Engineering for Transcriptional Control
Fundamental Principles and Quantitative Characterization
Core promoters are minimal DNA sequences that enable the formation of the transcription initiation complex and drive basal levels of transcription. In inducible systems, they are typically coupled with specific transcription factor binding sites (response elements) where the core promoter dictates the overall transcriptional efficiency while the response elements provide signal specificity [44]. Systematic quantitative characterization of core promoters is essential for selecting appropriate components for biosensor design.
A comprehensive study evaluating eight different core promoters demonstrated that choice of promoter significantly affects both basal expression levels and inducibility [44]. The research revealed that commonly used tried-and-true promoters like minimal CMV (minCMV) and minimal SV40 (minSV40), while providing robust gene expression upon induction, often suffer from high basal expression levels (leakiness), thereby reducing the fold-induction ratio [44]. Quantitative measurements showed that the panel of tested promoters spanned two to three orders of magnitude in basal gene-expression output, providing a wide selection for tuning synthetic systems [44].
Table 1: Quantitative Characterization of Core Promoter Performance in Mammalian Cells
| Core Promoter | Origin | Relative Basal Expression (%) | Fold-Induction | Key Characteristics |
|---|---|---|---|---|
| minCMV | Viral | >15% | Relatively small | Highest absolute induced expression; significant leakiness |
| minSV40 | Viral | Moderate | Moderate | Robust expression; moderate leakiness |
| CMV53 | Viral (minCMV derivative) | High | Moderate | Improved version with upstream GC box |
| miniTK | Viral | Low to moderate | Moderate | Herpes simplex thymidine kinase derivative |
| MLP | Viral | Low to moderate | Moderate | Adenovirus major late promoter |
| pJB42CAT5 | Mammalian (human junB) | Low | High | Reportedly robust, inducible activity |
| YB_TATA | Synthetic | Low | Significantly higher | Optimal combination of low basal expression with high transcription rate when induced |
| TATA box alone | Synthetic | Very low | Variable | Minimal promoter functionality |
Case Study: Engineering the Nar Promoter Library in E. coli
Promoter engineering has proven to be an efficient strategy for fine-tuning transcriptional control across various host organisms, including Escherichia coli, Corynebacterium glutamicum, Bacillus subtilis, and yeasts [45]. A representative example involves the engineering of a dissolved oxygen-dependent nar promoter in E. coli for fine-tuning expression levels of metabolic pathway genes.
Experimental Protocol: Construction and Screening of Synthetic Nar Promoter Library
- Library Construction: The 15-bp spacer region sequence between the consensus -35 and -10 elements of the wild-type
narpromoter was randomized using degenerated primers [45]. - Reporter Fusion: The randomized promoter fragments were fused via PCR to a DNA fragment consisting of a Shine-Dalgarno sequence, spacer, and His6-tagged GFPm (a reporter protein) [45].
- Cloning: The constructed promoter-reporter fragments were ligated into a pSTVM plasmid and transduced into E. coli TOP10 cells [45].
- High-Throughput Screening: The library (4.59 × 10^10 cells) was sorted by Fluorescence-Activated Cell Sorting (FACS) into three groups (low, intermediate, and high strength) through three rounds of sorting [45].
- Validation: Selected promoters were characterized through transcriptional analysis (qRT-PCR), protein expression (western blotting), and fluorescence intensity measurements [45].
This engineering approach generated synthetic nar promoters with significantly different strengths. The strong synthetic nar promoter (S3-2-64) and the intermediate promoter (W2U-30) showed 19.8 and 6.2 times higher strength, respectively, than the wild-type nar promoter based on fluorescence measurements, while the weak promoter (W2L-29) was 1.8 times weaker [45]. Sequencing revealed that strong promoters tended to have AT-rich spacers (33.3% GC for the strong promoter vs. 60% for wild-type), which enhance DNA flexibility and bendability [45].
Application in Metabolic Pathway Optimization:
The application of these tuned promoters significantly improved metabolic pathway yields. In a d-lactate pathway (single enzyme), using the strong synthetic nar promoter increased d-lactate titers by 34% (105.6 g/L vs. 79.0 g/L with wild-type) in fed-batch cultures [45]. More notably, in a three-gene 2,3-butanediol (BDO) pathway, the combinatorial optimization of promoter strengths (strong-weak-strong configuration) for the three genes increased BDO titers by 72% (88.0 g/L vs. 51.1 g/L with wild-type) [45], demonstrating the power of promoter fine-tuning in multi-gene pathways.
RBS Engineering for Translation Optimization
RBS Engineering Principles and Applications
While promoter engineering controls transcriptional initiation, RBS engineering provides a complementary approach for fine-tuning gene expression at the translational level. The RBS, located upstream of the start codon, plays a crucial role in recruiting ribosomes and initiating translation. In prokaryotic systems, graded gene expression can be effectively achieved through varying the strength of the RBS [44]. Key engineering strategies include:
- Sequence Modification: Altering the Shine-Dalgarno sequence and its spacing from the start codon to modulate translation initiation rates.
- Structural Accessibility: Engineering mRNA secondary structures that affect ribosomal binding accessibility.
- Library Approaches: Creating degenerate RBS libraries combined with high-throughput screening.
RBS engineering is particularly valuable for optimizing multi-gene pathways where stoichiometric balance between enzymes is critical for flux optimization. When combined with promoter engineering, RBS tuning enables multi-layered control strategies that separately address transcriptional and translational regulation.
Integration with Transcription Factor-Based Biosensors
Biosensor Architecture and Performance Metrics
Transcription factor-based biosensors are genetic devices that incorporate allosteric transcription factors (aTFs) as sensing components. These aTFs undergo conformational changes upon binding specific ligands (effectors), leading to activation or repression of downstream gene expression [10]. The core promoter and RBS elements directly determine the input-output relationship of these biosensors, influencing critical performance parameters:
Table 2: Key Biosensor Performance Parameters and Their Dependence on Genetic Components
| Performance Parameter | Definition | Influencing Genetic Components |
|---|---|---|
| Dynamic Range | Ratio between maximal and minimal output signal | Core promoter strength, RBS efficiency |
| Operating Range | Concentration window of optimal biosensor performance | TF-DNA binding affinity, promoter-RBS combination |
| Response Time | Speed of biosensor output change after stimulus | Promoter activation kinetics, protein maturation |
| Signal-to-Noise Ratio | Ratio between desired signal and background noise | Basal promoter activity, RBS leakage |
| Sensitivity | Lowest ligand concentration producing detectable response | TF-ligand binding affinity, promoter-RBS sensitivity |
| Specificity | Ability to distinguish target ligand from similar molecules | TF binding pocket, genetic context |
Computational Approaches for Biosensor Optimization
Recent advances in computational tools have significantly accelerated the engineering of biosensor components. Machine learning approaches like DeepTFactor predict novel transcription factors from genomic data [10] [9], expanding the available toolbox for biosensor design. For ligand specificity engineering, molecular docking and molecular dynamics simulations enable targeted alteration of TF ligand specificity through rational protein engineering [46].
A notable case study demonstrated the engineering of a LysR family BenM transcription factor to alter its specificity from its cognate ligand cis,cis-muconic acid to adipic acid through a single amino acid substitution identified via computational docking [46]. When implemented in a cell-free system, the engineered biosensor showed higher ligand sensitivity, highlighting the synergy between computational prediction and experimental implementation.
Experimental Protocols for Genetic Component Characterization
Quantitative Promoter Characterization Protocol
Objective: Quantitatively evaluate the performance of core promoters in mammalian cells.
Materials:
- Panel of core promoter constructs (e.g., minCMV, minSV40, YB_TATA, etc.)
- Reporter genes (e.g., Gaussia luciferase [Gluc], superfolder GFP [sfGFP])
- Normalization vector (e.g., constitutive CMV promoter driving dsRed-Express)
- Host cells (e.g., HEK 293T)
- Transfection reagents
- Measurement instruments (luminometer, flow cytometer)
Methodology:
- Clone each core promoter upstream of reporter genes (Gluc or sfGFP) in the absence of any response elements [44].
- Co-transfect with normalization vector (constitutive CMV-dsRed) to identify transfected cells and normalize for transfection efficiency [44].
- Measure basal gene expression levels using appropriate assays:
- Luciferase activity assays for Gluc
- Flow cytometry for sfGFP fluorescence intensity [44]
- Analyze data for:
- Relative basal expression levels across promoters
- Population distribution of expression (percentage of expressing cells)
- Fold-induction when coupled with response elements
Key Analysis: Compare both population-level expression (percentage of transfected cells showing detectable expression) and intensity-level expression (median fluorescence intensity among expressing cells) to fully characterize promoter performance [44].
High-Throughput Screening Protocol for Engineered Components
Objective: Identify optimized promoter or RBS variants from combinatorial libraries.
Materials:
- Combinatorial library of genetic variants
- Fluorescent reporter (e.g., GFP, RFP)
- Flow cytometer with cell sorting capability
- Microtiter plates for culture growth
Methodology:
- Transform variant library into host organism.
- Grow under selective conditions.
- Sort cells based on fluorescence intensity into subpopulations (e.g., low, intermediate, high) using multiple rounds of FACS [45].
- Isolate and sequence individual clones from each population.
- Characterize selected variants using secondary assays (e.g., qRT-PCR, western blotting) [45].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for Promoter and RBS Engineering
| Reagent / Tool | Function | Application Example |
|---|---|---|
| Degenerate Oligonucleotides | Library generation with randomized sequences | Randomizing promoter spacer regions or RBS sequences [45] |
| Fluorescent Reporters (GFP, RFP) | Quantitative measurement of gene expression | Screening promoter strength via flow cytometry [44] [45] |
| Enzymatic Reporters (Luciferase) | Highly sensitive quantification of expression | Precise measurement of promoter activity [44] |
| Flow Cytometer with Cell Sorter | High-throughput analysis and separation of cells | Sorting promoter/RBS library variants based on fluorescence [45] |
| Dual-Reporter Systems | Normalization for transfection efficiency | Constitutively expressed reference fluorophore (e.g., dsRed) [44] |
| qRT-PCR Reagents | Direct quantification of transcription levels | Validating transcriptional activity independent of translation [45] |
| Computational Prediction Tools | In silico design and optimization | DeepTFactor for TF prediction; docking for ligand specificity [10] [46] |
The fine-tuning of genetic components—specifically promoters and RBSs—represents a cornerstone in the development of high-performance transcription factor-based biosensors for dynamic regulation. Through quantitative characterization of component properties, strategic engineering of regulatory elements, and integration of computational design tools, researchers can create genetic circuits with precisely customized expression profiles. The continued refinement of these engineering approaches, coupled with advanced high-throughput screening methodologies, will undoubtedly expand the capabilities of biosensor technology, enabling more sophisticated applications in metabolic engineering, therapeutic development, and diagnostic systems. As the field progresses, the integration of promoter and RBS engineering with other regulatory layers will provide unprecedented control over cellular behavior, paving the way for next-generation synthetic biology applications.
Transcription factor-based biosensors (TFBs) are powerful synthetic biology tools that convert the presence of a specific metabolite into a quantifiable genetic output, enabling real-time monitoring of intracellular metabolites, dynamic control of metabolic pathways, and high-throughput screening for strain engineering [1] [9]. While native TF systems provide a starting point, their performance is often hampered by limited specificity, dynamic range, and cross-reactivity with host regulatory networks. The emerging integration of computational tools has begun to systematically address these limitations, creating a new paradigm for the rational design of sophisticated biosensing systems [1] [46].
This technical guide examines three pivotal computational technologies transforming TFB development: Cello for genetic circuit design and optimization, DeepTFactor for predicting transcription factors from protein sequences, and Molecular Docking for engineering ligand specificity. These tools enable researchers to move beyond traditional trial-and-error approaches, offering predictive power and precision that significantly accelerate the development of bespoke biosensors for dynamic regulation research, metabolic engineering, and therapeutic development [46] [47]. The integration of these computational approaches provides a comprehensive workflow from transcription factor discovery to the implementation of optimized genetic circuits, ultimately enhancing our ability to program cellular behavior for scientific and industrial applications.
Computational Foundations for Next-Generation Biosensors
Cello: Automated Genetic Circuit Design
Cello is a genetic design automation tool that applies principles from electronic circuit design to synthetic biology, enabling the in silico design and optimization of complex genetic circuits before physical implementation [1]. This software uses a hardware description language (Verilog) to abstract biological components into standardized logic gates, allowing researchers to specify desired circuit behavior without needing to manually design every molecular component. Cello then converts these logical specifications into DNA sequences that implement the desired function within microbial hosts, significantly reducing the design-build-test cycle time for sophisticated genetic circuits [1].
Within the context of transcription factor-based biosensors, Cello provides particular value for integrating multiple TF inputs into circuits that perform logical operations, enabling more sophisticated sensing and response capabilities. For instance, researchers can design circuits that only activate reporter genes or metabolic pathways when specific combinations of metabolites are present, creating precision control systems that minimize off-target effects in dynamic regulation experiments [1]. The software's ability to predict circuit performance under different growth conditions and its extensive library of characterized genetic parts further enhances its utility for developing reliable biosensing systems for research and bioproduction applications.
Table 1: Key Features and Applications of Cello in Biosensor Development
| Feature | Technical Description | Application in TFB Research |
|---|---|---|
| Verilog Compiler | Translates logic operations into DNA sequences | Creates complex genetic circuits from simple code |
| Parts Library | Repository of characterized promoters, RBSs, terminators | Ensures reliable part performance in biosensor design |
| Performance Prediction | Models circuit behavior under different conditions | Optimizes biosensor dynamic range and sensitivity |
| UCF (User Constraint File) | Defines host-specific operating conditions | Tailors biosensors for specific microbial chassis |
| Logic Gate Integration | Combines multiple TF inputs into single outputs | Enables multi-analyte sensing and decision-making |
DeepTFactor: High-Accuracy Transcription Factor Prediction
DeepTFactor represents a significant advancement in bioinformatics tools for transcription factor identification. This deep learning-based method employs a convolutional neural network trained on the largest available dataset of TFs and non-TFs to predict whether a given protein sequence functions as a transcription factor [47]. The tool achieves an impressive Area Under the Curve (AUC) of 0.97 on independent test datasets, demonstrating high reliability for TF annotation in genomic and metagenomic studies [47].
For biosensor development, DeepTFactor addresses a fundamental challenge: the limited repertoire of well-characterized transcription factors specific to molecules of interest. Before engineering can begin, researchers must first identify candidate TFs that might respond to their target analyte. DeepTFactor enables rapid screening of large protein datasets, including those derived from metagenomic studies of non-culturable organisms, significantly expanding the pool of potential biosensor components [47]. This capability is particularly valuable for developing biosensors for emerging contaminants or rare metabolites where few native sensing systems are documented.
Experimental Protocol: Implementing DeepTFactor for TF Discovery
Data Collection: Obtain protein sequences of interest from databases such as UniProtKB/Swiss-Prot. For metagenomic mining, assemble contigs and predict open reading frames using tools like Prodigal or MetaGeneMark.
Sequence Preprocessing: Remove redundant sequences and sequences containing non-natural amino acids to ensure data quality. The final dataset should be in FASTA format.
TF Prediction: Submit the preprocessed FASTA file to the DeepTFactor web server (available at https://webs.iiitd.edu.in/raghava/transfacpred) or run the standalone version locally. The tool will generate prediction scores for each sequence.
Result Interpretation: Sequences with prediction scores above 0.5 are classified as transcription factors. Higher scores indicate greater confidence in the prediction.
Validation: Confirm predictions through sequence alignment with known TF families (e.g., LysR, TetR, AraC) and experimental validation when possible.
The recent development of TransFacPred, a hybrid method that combines alignment-based and alignment-free approaches, has further enhanced TF prediction capabilities, achieving an AUC of 0.99 on independent datasets [47]. This integrated approach leverages the precision of homology-based methods with the broader coverage of machine learning-based techniques, providing comprehensive coverage for TF identification across diverse organisms.
Molecular Docking: Engineering Biosensor Specificity
Molecular docking computational techniques predict the binding affinity and orientation of ligands within protein binding pockets, making them invaluable for engineering transcription factor specificity [48] [46]. In the context of TFBs, docking simulations help researchers understand how TFs interact with their ligand effectors at an atomic level, enabling rational redesign of binding pockets to alter specificity, improve sensitivity, or create novel biosensors for non-native ligands [46].
The computational workflow typically involves several key steps: preparing the 3D structures of the TF and ligand, sampling possible binding conformations, and scoring these conformations based on their binding affinities. Advanced docking approaches can handle flexibility in both the ligand and protein, providing more accurate predictions of binding modes [48]. For biosensor engineering, this capability enables targeted alteration of TF ligand specificity through informed mutagenesis of key binding pocket residues.
Experimental Protocol: Specificity Engineering with Molecular Docking
Structure Preparation:
- Obtain the 3D structure of the transcription factor from the Protein Data Bank (PDB) or generate a homology model.
- Prepare the ligand structure in MOL2 or SDF format, optimizing its geometry using energy minimization.
Binding Site Identification:
- Define the binding pocket using literature data or computational prediction tools.
- Alternatively, use blind docking to scan the entire protein surface for potential binding sites.
Docking Simulation:
- Use docking software such as AutoDock Vina, AutoDock, or DOCK to generate multiple ligand poses within the binding site.
- Apply search algorithms (systematic, stochastic, or genetic algorithms) to explore possible binding conformations.
Pose Analysis and Scoring:
- Evaluate generated poses using scoring functions (force field-based, empirical, or knowledge-based).
- Identify hotspot residues critical for ligand binding and specificity.
Mutation Planning:
- Select candidate residues for mutagenesis based on their interactions with the ligand.
- Use the docking results to predict how specific mutations would alter ligand binding.
A compelling example of this approach comes from a study that engineered the LysR family BenM TF to alter its specificity from its cognate ligand cis,cis-muconic acid to adipic acid [46]. Through a computation-guided workflow utilizing molecular docking, researchers identified a single amino acid substitution that successfully switched ligand specificity. When implemented in a cell-free system, the engineered biosensor showed enhanced ligand sensitivity, demonstrating the practical utility of this computational approach for expanding the biosensor repertoire [46].
Integrated Workflow for Computational Biosensor Development
The true power of computational tools emerges when they are integrated into a cohesive workflow for biosensor development. This integrated approach combines the strengths of each tool while mitigating their individual limitations, creating a robust pipeline from transcription factor discovery to functional genetic circuit implementation.
Diagram 1: Integrated computational workflow for developing transcription factor-based biosensors, showing the sequential relationship between key tools and processes.
The workflow begins with Transcription Factor Prediction using tools like DeepTFactor or the hybrid TransFacPred method to identify candidate TFs from genomic or metagenomic data [47]. This step significantly expands the repertoire of potential biosensor components beyond well-characterized model organisms. Following identification, Structure Modeling and Preparation ensures high-quality 3D structures are available for subsequent computational analysis, either from experimental data or homology modeling.
The core engineering phase employs Molecular Docking to understand and alter ligand specificity by identifying key residues in the binding pocket that govern molecular recognition [46]. This computational guidance enables targeted mutagenesis to create TFs with customized specificity profiles. Finally, Genetic Circuit Design with Cello integrates the engineered TF into a functional biosensor circuit with optimized performance characteristics, properly insulated from host regulatory networks, and potentially combined with other sensing modules for complex logical operations [1].
This integrated computational approach dramatically reduces the experimental trial-and-error typically associated with biosensor development. For instance, a researcher aiming to develop a biosensor for a novel metabolite could use DeepTFactor to scan metagenomic databases from unique environmental niches, identify potential TF candidates, employ molecular docking to understand and potentially improve ligand binding, and finally use Cello to incorporate the optimized TF into a genetic circuit with the desired input-output relationship.
Research Reagent Solutions: Essential Tools for Computational Biosensor Development
Table 2: Key Research Reagents and Computational Tools for Biosensor Development
| Tool/Reagent | Type | Primary Function | Application Notes |
|---|---|---|---|
| Cello | Software | Genetic circuit design automation | Converts Verilog code to DNA sequences; requires User Constraint Files for specific hosts |
| DeepTFactor | Web Server/Standalone | Transcription factor prediction | Uses deep learning; AUC of 0.97 on independent datasets |
| TransFacPred | Hybrid Prediction Tool | TF identification | Combines alignment-based and alignment-free methods; AUC of 0.99 |
| AutoDock Vina | Molecular Docking Software | Protein-ligand docking prediction | Open-source; uses machine learning-based scoring functions |
| AutoDock | Molecular Docking Suite | Flexible ligand docking | Lamarckian genetic algorithm for pose optimization |
| UCSF DOCK | Docking Software | Structure-based drug design | Grid-based method for binding affinity evaluation |
| JASPAR | Database | Curated transcription factor binding profiles | Source of position frequency matrices for binding sites |
| RegulonDB | Database | E. coli transcriptional regulation | Reference for known regulatory networks in E. coli |
| SM-TF Database | Database | Small molecule-TF complex structures | Source of 3D structures for docking studies |
| Cell-Free Systems | Experimental Platform | Biosensor characterization | Enables sensitive testing without cellular context limitations |
The integration of computational tools like Cello, DeepTFactor, and molecular docking represents a paradigm shift in transcription factor-based biosensor development. These technologies enable a more predictive, rational design approach that significantly accelerates the creation of sophisticated biosensing systems for dynamic regulation research [1] [46] [47]. As these tools continue to evolve, we can anticipate several exciting developments that will further enhance their capabilities and applications.
Future advancements will likely include tighter integration between these tools, creating seamless workflows from TF discovery to circuit implementation. We can also expect improved accuracy in molecular docking through better handling of protein flexibility and more sophisticated scoring functions [48]. The integration of molecular dynamics simulations with docking studies will provide deeper insights into the structural dynamics responsible for ligand recognition and allosteric control [46]. Additionally, the expanding application of artificial intelligence and machine learning across all aspects of biosensor design promises to further reduce development timelines and expand the range of detectable analytes [1] [49].
For researchers in dynamic regulation and metabolic engineering, these computational tools offer unprecedented capability to design bespoke biosensors that precisely monitor and control metabolic fluxes in real-time. This capability is particularly valuable for optimizing microbial cell factories for bioproduction, where dynamic regulation can help balance growth and production phases, redirect metabolic fluxes, and improve overall titers and yields [9]. As the repertoire of computational tools continues to expand and their integration becomes more seamless, we can anticipate a new era of precision biosensor design that will transform both fundamental research and industrial applications in synthetic biology.
The engineering of transcription factors (TFs) represents a frontier in synthetic biology, enabling the construction of sophisticated biosensors for dynamic metabolic regulation. Traditional genetic circuits have been constrained by the limited repertoire of natural transcriptional regulators, each possessing a fixed input-output connection. To overcome this fundamental limitation, two powerful protein engineering paradigms have emerged: domain swapping, which recombines existing functional modules from natural proteins, and de novo protein design, which creates entirely new proteins from first principles. Framed within the context of developing advanced transcription factor-based biosensors, these approaches collectively expand the synthetic biology toolkit, allowing researchers to engineer novel genetic connections and programmable cellular behaviors previously inaccessible through natural parts alone. This technical guide examines the core principles, methodologies, and applications of these complementary strategies, providing researchers with the experimental frameworks needed to implement these advanced techniques in biosensor development for metabolic engineering and therapeutic applications.
Domain Swapping: Engineering Modularity through Recombination
Core Principles and Strategic Framework
Domain swapping is a protein engineering strategy that creates hybrid transcriptional regulators by exchanging discrete functional domains between homologous proteins within the same family. The fundamental premise is that many transcriptional regulators are composed of modular domains—typically a DNA-binding domain (DBD) that recognizes specific promoter sequences and a signal-sensing domain (SDD) that responds to effector molecules—that can be functionally interchanged [50]. This strategy directly addresses a major constraint in synthetic biology: the inability to flexibly connect biological signals to genetic outputs using natural regulators that possess fixed input-output specificities [50].
The strategic implementation of domain swapping follows a systematic workflow that begins with the identification of target protein families exhibiting suitable modular architecture. Successful applications have been demonstrated across numerous regulator families, including LacI, TetR, LuxR, OmpR/NarL, and MerR [50]. The key to success lies in selecting proteins with conserved domain boundaries and high structural similarity at the fusion points. Structural data, particularly from X-ray crystallography, is invaluable for identifying discrete, functional, and conserved DBDs and SDDs [50]. For many bacterial regulators, the DBD typically contains a helix-turn-helix (HTH) motif that governs DNA recognition and binding, while the SDD undergoes conformational changes upon ligand binding [50].
Experimental Methodology and Implementation
The domain swapping workflow encompasses both computational and experimental phases, with careful validation at each stage. Below is the detailed experimental protocol for creating hybrid regulators through domain swapping.
Table 1: Key Stages in Domain Swapping Experimental Protocol
| Stage | Key Activities | Output |
|---|---|---|
| 1. Target Identification | Select homologous TF families with modular architecture; analyze available structural data (X-ray, NMR, AF2) | Candidate protein pairs with defined domain boundaries |
| 2. In Silico Design | Identify conserved fusion points; generate hybrid sequences; model chimeric structures | Library of hybrid protein sequences |
| 3. Genetic Construction | Use overlap extension PCR or synthetic gene synthesis; clone into expression vectors | Plasmid library encoding hybrid regulators |
| 4. Functional Screening | Transform into host chassis; measure reporter expression with/without ligand | Primary functional hits |
| 5. Characterization | Determine dose-response curves, dynamic range, specificity, potential cross-talk | Quantitative performance metrics |
| 6. Engineering & Optimization | Apply directed evolution to problematic hybrids; rational mutagenesis | Optimized biosensor components |
Phase 1: Template Identification and Analysis
- Select transcription factor families with known modular architecture and available structural information
- Identify domain boundaries by analyzing conserved regions across multiple sequence alignments and structural data
- Prioritize fusion points in regions with high sequence conservation and structural flexibility, typically in inter-domain linkers
Phase 2: Hybrid Design and Construction
- Design hybrid genes using overlap extension PCR or synthetic gene synthesis
- For overlap extension PCR: Design primers with overlapping regions between DBD of one regulator and SDD of another
- Clone constructed hybrids into appropriate expression vectors with selectable markers
- Include control vectors expressing parent regulators for comparative analysis
Phase 3: Functional Validation and Characterization
- Transform hybrid constructs into microbial chassis (typically E. coli)
- Measure reporter gene expression (e.g., fluorescence) across a range of ligand concentrations
- Calculate dynamic range (fold-change between induced and uninduced states) and EC50/IC50 values
- Assess specificity by challenging with structurally similar molecules to detect cross-reactivity
Phase 4: Optimization and Troubleshooting Addressing non-functional hybrids remains a challenge in domain swapping, with studies reporting approximately 50% of initial constructs may be poorly functional [50]. Optimization strategies include:
- Computational rescue: Use sequence analysis and structural modeling to identify and rectify problematic regions [50]
- Directed evolution: Implement iterative mutagenesis and screening to restore function to compromised hybrids
- Rational mutagenesis: Introduce targeted mutations at critical positions based on structural insights
Applications in Genetic Circuit Design
Domain-swapped modular regulators enable sophisticated genetic circuit topologies that were previously infeasible. Notable implementations include:
Multi-input Logic Gates: By using multiple regulators that recognize the same promoter but respond to different signals, researchers have created AND gates where all corresponding signals must coexist to activate reporter expression [50].
Layered Circuitry: Groseclose et al. produced a cascade circuit where one hybrid regulator controls the expression of another, creating a temporal program in genetic circuits [50].
Dynamic Metabolic Control: Modular regulators have been implemented in metabolic engineering to dynamically control flux distribution, enabling automatic redirection of resources between growth and production phases [3].
The quantitative performance of domain-swapped biosensors can be enhanced through systematic engineering. For instance, in a CaiF-based biosensor for L-carnitine, researchers employed a "Functional Diversity-Oriented Volume-Conservative Substitution Strategy" to create variant CaiF(Y47W/R89A) that exhibited a 1000-fold wider concentration response range (10â»â´ mM to 10 mM) and a 3.3-fold higher output signal intensity compared to the wild-type biosensor [36].
De Novo Protein Design: Creating Novel Proteins from First Principles
Fundamental Concepts and Paradigm Shift
De novo protein design represents a paradigm shift from engineering natural proteins to creating entirely novel proteins with customized folds and functions not observed in nature. This approach aims to transcend the limitations of natural evolutionary history, which has produced proteins optimized for biological fitness rather than human utility [51]. The core premise is that the known natural fold space represents only a tiny fraction of the theoretically possible protein functional universe, with recent evidence suggesting natural fold discovery is approaching saturation [51].
The de novo design process is governed by the sequence → structure → function paradigm—the principle that a protein's amino acid sequence encodes its three-dimensional fold, which in turn determines its biological function [51]. The challenge lies in the astronomical scale of sequence space: a mere 100-residue protein theoretically permits 20¹â°â° (≈1.27 × 10¹³â°) possible amino acid arrangements, exceeding the estimated number of atoms in the observable universe by more than fifty orders of magnitude [51]. Navigating this vast space to find sequences that fold into stable structures and perform useful functions requires sophisticated computational approaches beyond traditional methods.
Computational Frameworks and Design Strategies
Modern de novo protein design has been revolutionized by artificial intelligence (AI) and machine learning, which complement and extend traditional physics-based design methods. The table below compares major computational approaches.
Table 2: Computational Methods for De Novo Protein Design
| Method | Core Approach | Key Applications | Advantages | Limitations |
|---|---|---|---|---|
| Rosetta | Physics-based energy minimization; fragment assembly | Top7 novel fold; enzyme active sites; binding scaffolds | Proven versatility; detailed atomic modeling | Computationally expensive; force field approximations |
| RFdiffusion | Generative diffusion models; structure conditioning | De novo homodimeric TFs; DBP backbone generation | Explores novel folds; high design success rate | Requires specialized computational resources |
| ProteinMPNN | Neural network-based sequence design | Amino acid sequence generation from backbone structures | Fast; high-quality sequences | Dependent on accurate backbone input |
| AlphaFold2 | Deep learning structure prediction | Structure validation; scaffold library generation | High prediction accuracy; rapid modeling | Limited to natural-like structures |
RFdiffusion and ProteinMPNN Pipeline for Transcription Factor Design: Recent breakthroughs have demonstrated the feasibility of designing de novo DNA-binding proteins (DBPs) and transcription factors through a structured computational pipeline [52] [53]. The methodology involves:
- Scaffold Generation: Using RFdiffusion to design homodimeric TF backbones optimized for DNA recognition [52]
- Sequence Design: Applying ProteinMPNN to generate amino acid sequences that stabilize the designed backbone [53]
- Structure Prediction: Validating designs with AlphaFold2 to assess folding confidence (pLDDT) and structural accuracy [53]
- Experimental Validation: Expressing designed TFs in E. coli and measuring repression through fluorescence assays [52]
This approach has yielded six de novo TFs achieving over 4-fold repression, with the highest performer reaching nearly 20-fold repression—comparable to CRISPR interference systems [52]. The designs demonstrated considerable orthogonality, highlighting the precision of computational methods.
Key Design Principles for DNA-Binding Proteins: Successful de novo design of DBPs requires addressing several fundamental challenges [53]:
- Backbone Positioning: Strategic use of main-chain phosphate hydrogen bonds to precisely position scaffolds for base contacts
- Specific Base Recognition: Focus on polar interactions with nucleotide base atoms in the major groove
- Side Chain Preorganization: Engineering hydrogen-bond networks to rigidly position long polar side chains for specific base recognition
The RIFdock algorithm addresses these challenges by comprehensively sampling disembodied side-chain interactions with DNA base atoms, then finding scaffold placements that satisfy both backbone hydrogen bonds and base-contacting side chains [53].
Implementation and Experimental Validation
Protocol for De Novo TF Design and Testing:
Phase 1: Computational Design
- Define target DNA sequence and generate initial scaffold backbones using RFdiffusion
- Focus on helix-turn-helix (HTH) domains as they are compact and capable of major groove interactions
- Generate amino acid sequences using ProteinMPNN with DNA atoms incorporated in the interaction graph
- Filter designs based on predicted local distance difference test (pLDDT), predicted alignment error (PAE), and structural metrics
Phase 2: Experimental Characterization
- Synthesize genes encoding top designs (typically 96 candidates) and clone into expression vectors
- Transform into E. coli reporter strains with fluorescent proteins under control of target promoters
- Measure repression capability through fluorescence assays comparing expression with and without TF expression
- Assess orthogonality by testing cross-reactivity with non-cognate DNA sequences
Phase 3: Optimization and Application
- For successful designs, further optimize through computational inpainting to resample scaffold loops
- Integrate designed TFs into genetic circuits for biosensing applications
- Characterize performance in dynamic regulation scenarios relevant to metabolic engineering
This approach has proven successful, with designed DBPs functioning in both E. coli and mammalian cells to repress and activate transcription of neighboring genes [53]. The method provides a route to small, readily deliverable sequence-specific DBPs for sophisticated gene regulation applications.
The Scientist's Toolkit: Essential Research Reagents and Materials
Implementation of domain swapping and de novo design approaches requires specialized reagents and computational resources. The following table catalogues essential materials for researchers embarking on these protein engineering strategies.
Table 3: Essential Research Reagents and Resources for Advanced Protein Engineering
| Category | Specific Reagents/Resources | Function/Application | Key Considerations |
|---|---|---|---|
| DNA Assembly | Overlap extension PCR reagents; synthetic gene fragments | Hybrid gene construction; de novo gene synthesis | Quality of synthetic genes critical for success |
| Expression Systems | Modular plasmid vectors; inducible promoters | Heterologous expression in microbial chassis | Promoter strength affects TF performance |
| Reporter Systems | Fluorescent proteins (GFP, RFP); luciferase | Quantitative measurement of TF activity | Dynamic range and sensitivity requirements |
| Screening Platforms | Flow cytometers; microplate readers | High-throughput functional characterization | Throughput capabilities affect library size |
| Computational Tools | Rosetta; RFdiffusion; ProteinMPNN; AlphaFold2 | In silico design and validation | Computational resource requirements vary |
| Structural Data | Protein Data Bank entries; AlphaFold DB | Template identification; fusion point design | Quality and resolution impact design accuracy |
| Directed Evolution | Mutagenesis kits; FACS equipment | Optimization of poorly functional hybrids | Library size and screening efficiency critical |
Comparative Analysis and Strategic Implementation
Technical Comparison and Selection Guidelines
Domain swapping and de novo design offer complementary advantages for transcription factor engineering. The table below compares their key characteristics to guide selection based on project requirements.
Table 4: Domain Swapping vs. De Novo Design: Comparative Analysis
| Parameter | Domain Swapping | De Novo Design |
|---|---|---|
| Prerequisites | Requires homologous natural templates with modular architecture | Requires only target DNA sequence and computational resources |
| Development Timeline | Medium (weeks to months) | Long (months to years) |
| Success Rate | Moderate (~50% initial functionality) [50] | Low but improving with AI methods |
| Performance Level | Moderate to high (builds on optimized natural domains) | Variable (4-20 fold repression demonstrated) [52] |
| Technical Barrier | Medium (requires molecular biology expertise) | High (requires computational expertise and resources) |
| Key Advantages | Builds on proven natural folds; predictable outcomes | Access to novel specificities beyond natural constraints |
| Primary Limitations | Constrained by natural sequence space; potential hybrid dysfunction | Uncertain folding in vivo; limited predictability of function |
Implementation Roadmap for Biosensor Development
Strategic implementation of these technologies for transcription factor-based biosensors follows a structured approach:
Phase 1: Project Scoping and Requirements Analysis
- Define target analyte and required detection parameters (sensitivity, dynamic range, specificity)
- Assess availability of natural TFs for target or related compounds
- Evaluate computational resources and expertise available
Phase 2: Technology Selection
- Domain swapping preferred when: Natural TF families with target specificity exist; project timeline constrained; experimental resources limited
- De novo design preferred when: No natural TFs available for target; novel specificities required; computational resources substantial
Phase 3: Iterative Design-Build-Test Cycles
- Implement appropriate workflow (domain swapping or de novo)
- Incorporate high-throughput screening for functional characterization
- Apply optimization strategies (directed evolution, computational rescue) to initial designs
Phase 4: Integration and Deployment
- Incorporate functional biosensors into genetic circuits for metabolic engineering
- Implement dynamic control strategies for pathway optimization
- Validate performance in real-world application contexts
Future Perspectives and Emerging Trends
The convergence of domain swapping and de novo design with artificial intelligence represents the future of transcription factor engineering. Several emerging trends are particularly noteworthy:
AI-Accelerated Design: Deep learning approaches are rapidly advancing, with tools like RoseTTAFold2NA and DeepFoldRNA improving our ability to model nucleic acid/protein complexes [10]. These developments will enhance both domain boundary prediction for swapping and structure prediction for de novo design.
Hybrid Approaches: Future workflows will likely combine the best aspects of both strategies—using de novo design to create novel DNA-binding specificities, then applying domain swapping principles to integrate these with optimized sensing domains from natural proteins.
Expanded Application Space: As these technologies mature, applications will grow beyond metabolic engineering to include diagnostic biosensors, therapeutic regulatory circuits, and environmental monitoring systems [54] [8].
The integration of these advanced protein engineering approaches with high-throughput characterization and machine learning-guided optimization promises to dramatically accelerate the development of transcription factor-based biosensors, ultimately enabling sophisticated dynamic regulation schemes that bring synthetic biology closer to realizing its full potential in biotechnology and medicine.
Ensuring Efficacy: Validation Frameworks and Comparative Technology Analysis
Transcription factor (TF)-based biosensors are powerful synthetic biology tools that convert the intracellular presence of a specific ligand into a quantifiable output, typically gene expression [10] [9]. Their application is pivotal for dynamic regulation of metabolic pathways, high-throughput screening of production strains, and real-time monitoring of metabolites [14] [8]. However, the journey from a conceptual biosensor design to a functionally reliable tool is often hindered by performance issues such as non-specific activity, leaky expression, and cross-talk with host cellular networks [10] [2]. A structured validation workflow that seamlessly connects in silico predictions with experimental prototyping is therefore essential to overcome these challenges and accelerate the development of robust, application-ready biosensors. This guide provides a detailed technical framework for such a workflow, specifically contextualized within TF-based biosensor research for dynamic regulation.
Phase I: In Silico Design and Computational Analysis
The initial phase focuses on computational design and prediction to generate high-quality biosensor designs before any wet-lab experiments, saving significant time and resources.
Knowledge Base and TF Selection
The foundation of a robust biosensor is a well-characterized allosteric transcription factor (aTF). The following databases are critical for the initial discovery and selection process [10] [9].
Table 1: Key Databases for Transcription Factor and Biosensor Data
| Database Name | Primary Focus | Application in Workflow |
|---|---|---|
| RegulonDB [9] | Transcriptional regulation in E. coli K-12 | Reference for well-characterized aTFs in a model organism. |
| P2TF [9] | Predicted Prokaryotic TFs | Compilation of TFs from sequenced genomes and metagenomes. |
| JASPAR [9] | Curated TF binding profiles (PFMs) | Identification of DNA binding motifs for biosensor output modules. |
| SM-TF [9] | 3D structures of small molecule-TF complexes | Source of structural data for computational modeling and engineering. |
| Bionemo [9] | Proteins and genes in biodegradation metabolism | Finding aTFs responsive to environmental pollutants or related metabolites. |
Computational Modeling and Simulation
With a candidate aTF identified, in silico modeling predicts biosensor behavior and informs engineering strategies.
- Genetic Circuit Modeling: Software tools like Cello enable the in silico design and simulation of genetic circuits, allowing for the prediction of output dynamics based on promoter strength, RBS tuning, and TF-DNA binding kinetics [55] [8]. This helps in selecting circuit architectures with the desired dynamic range and sensitivity before construction.
- Structure-Guided Engineering: When an aTF requires optimization for a new ligand, computational tools are indispensable. As demonstrated in the engineering of BenM for adipic acid detection, a workflow involving molecular docking to identify key residues in the ligand-binding pocket, followed by site-directed mutagenesis, can successfully alter ligand specificity [46]. Molecular dynamics simulations can further analyze how mutations affect TF structure and binding mechanics [46].
- AI-Enhanced Prediction: Machine learning tools like DeepTFactor can predict novel TFs from genomic sequences, expanding the repertoire of available sensing components [10] [9]. Furthermore, protein structure prediction tools like AlphaFold can generate reliable 3D models of aTFs, providing structural data for docking studies even when experimental structures are unavailable [10].
Figure 1: In Silico Design Workflow for TF-Based Biosensors
Phase II: High-Throughput Experimental Prototyping
The computationally designed biosensor variants must be rapidly tested and characterized. This phase leverages cell-free systems and automated liquid handling for high-throughput experimental validation.
Cell-Free Gene Expression (CFE) for Rapid Screening
Cell-free systems bypass the need for time-consuming cell transformation and culture, enabling direct testing of biosensor circuits encoded in linear DNA templates [56]. A key application is the rapid screening of aTF mutant libraries for desired ligand specificity and sensitivity [56] [46].
Table 2: Key Reagents and Parameters for Cell-Free Biosensor Characterization
| Reagent / Parameter | Function / Role | Considerations for Validation |
|---|---|---|
| Linear Expression Template (LET) | DNA template encoding the biosensor circuit; produced via PCR. | Circumvents plasmid cloning; enables rapid testing of thousands of designs [56]. |
| Cellular Extract | Provides transcriptional and translational machinery. | Batch-to-batch consistency is critical for reproducible results [56]. |
| Ligand Stock Solutions | Used to challenge the biosensor and generate a dose-response. | Purity and accurate concentration are vital for determining sensitivity and selectivity [56] [46]. |
| Fluorescent Reporter (e.g., sfGFP) | Quantifiable output for biosensor activation. | Enables high-throughput measurement using plate readers [56]. |
| Reaction Volume & Miniaturization | Conducting assays in 1-10 µL volumes in 384-well plates. | Reduces reagent costs and enables massive parallelism [56]. |
| Liquid Handler Transfer Settings | Automated nanoliter-volume dispensing of reagents. | Fluid properties (viscosity, surface tension) must be calibrated (e.g., using B2 preset on Echo 550) for accuracy [56]. |
Automated Workflow and Quality Control
Implementing an automated CFE workflow requires rigorous validation of the liquid handling process itself to ensure data quality [56].
- Fluid Transfer Optimization: The acoustic liquid handler (e.g., Echo) must be configured with the correct fluidic preset (e.g., "B2" for DNA in PCR buffer) to ensure precise and consistent droplet transfer [56].
- Assay Quality Metrics: The Z'-factor should be calculated to validate the assay's robustness for high-throughput screening. A Z' > 0.5 is considered acceptable, indicating a sufficient dynamic range between positive and negative controls [56].
- Plate Effect Mitigation: The experimental design should account for "edge effects" and material drift across the plate by using interleaved control patterns to identify and correct for these confounding artifacts [56].
Figure 2: High-Throughput Cell-Free Validation Workflow
Phase III: In Vivo Implementation and System-Level Validation
Biosensor candidates that perform well in cell-free systems must be validated within living cells, where they interact with the full complexity of the host's physiology.
Strain Engineering and In Vivo Characterization
The validated genetic construct is integrated into the host chassis, such as E. coli, for final testing and application.
- Dynamic Range and Operational Range: The biosensor's performance is characterized in vivo by measuring its output (e.g., fluorescence) across a range of ligand concentrations. The dynamic range is the fold-change between the "on" and "off" states, while the operational range is the concentration window over which it responds [36] [2]. These parameters can be tuned by engineering the aTF itself or by modifying genetic parts like promoters and ribosome-binding sites [2] [8].
- Directed Evolution for Performance Enhancement: If the biosensor's dynamic range is insufficient, directed evolution is a powerful strategy. As demonstrated with the CaiF biosensor for L-carnitine, a "Functional Diversity-Oriented" saturation mutagenesis strategy can generate variants with a 1000-fold wider response range and a 3.3-fold higher signal output [36].
- Application in Dynamic Regulation: The ultimate test of a biosensor for dynamic regulation is its integration into a control circuit. For example, the FapR-based malonyl-CoA biosensor in E. coli strain FA3 was used to dynamically regulate the expression of acetyl-CoA carboxylase (ACC), a key enzyme in fatty acid biosynthesis. This biosensor-enabled feedback controller prevented the cytotoxic overaccumulation of ACC, leading to improved fatty acid yields [57].
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Biosensor Validation
| Item / Reagent | Function in Workflow | Specific Example / Note |
|---|---|---|
| Acoustic Liquid Handler | For precise, nL-volume transfer of CFE reagents. | Echo 525/550 with optimized fluidic presets (e.g., B2 for DNA) [56]. |
| Cell-Free Protein Synthesis System | In vitro transcription/translation system for rapid biosensor testing. | Crude E. coli extract-based systems compatible with LETs [56]. |
| Fluorescent Reporter Proteins | Quantifiable output for biosensor activity. | superfolder GFP (sfGFP) is a common, stable choice [56]. |
| Model Host Organism | Chassis for in vivo validation and application. | Escherichia coli K-12 strains (e.g., MG1655) with extensive known regulons (RegulonDB) [9] [57]. |
| Directed Evolution Kit | For creating and screening mutant aTF libraries. | Kits for site-saturation mutagenesis (e.g., NNK codons) followed by FACS or microplate screening [36]. |
| Biosensor Plasmid Backbone | Standardized vector for in vivo expression. | Vectors with tunable promoters (e.g., PLtetO-1) and orthogonal reporter genes [2]. |
A rigorous, multi-phase validation workflow is non-negotiable for developing reliable transcription factor-based biosensors. By starting with comprehensive in silico design using curated databases and computational models, progressing to high-throughput experimental prototyping in cell-free systems, and culminating in robust in vivo testing and implementation, researchers can effectively de-risk the development process. This structured approach, leveraging the tools and methodologies outlined in this guide, enables the creation of high-performance biosensors capable of driving advances in dynamic metabolic engineering, high-throughput screening, and intelligent bioproduction.
Characterizing Dose-Response Curves and Performance in Relevant Hosts
Transcription factor (TF)-based biosensors are indispensable tools in synthetic biology, serving as the foundational components for dynamic regulation in metabolic engineering and high-throughput screening strategies. These genetically encoded devices function by converting the intracellular concentration of a specific small molecule (ligand) into a quantifiable gene expression output [8]. The dose-response curve, which graphically represents this input-output relationship, provides the most critical quantitative description of biosensor performance. A thorough characterization of this curve is not merely a procedural step but an absolute prerequisite for the effective deployment of biosensors in any application, especially for dynamic regulation within complex metabolic networks. Proper characterization reveals how the biosensor will behave under physiological conditions, informing researchers whether its sensitivity, range, and output are suited to the target pathway [6] [3]. Without this deep understanding, attempts at dynamic control are likely to fail due to mismatches between the biosensor's operational parameters and the host's metabolic reality.
This guide provides an in-depth technical framework for characterizing the dose-response curves of TF-based biosensors and evaluating their performance in relevant host chassis. It synthesizes current methodologies, performance metrics, and tuning strategies to equip researchers with the knowledge to rigorously validate and optimize these powerful tools for robust dynamic regulation.
Key Performance Parameters of Dose-Response Curves
The performance of a TF-based biosensor is quantified by several key parameters derived from its dose-response curve. These metrics collectively define the operational window and reliability of the biosensor within a host organism [6] [3].
- Dynamic Range is the fold-change between the minimal (uninduced) and maximal (induced) output signal. A large dynamic range is crucial for distinguishing between low and high producer strains in screening or generating a strong regulatory signal in dynamic control.
- Sensitivity (ECâ‚…â‚€ or Kâ‚) reflects the ligand concentration required to elicit a half-maximal response. It determines the biosensor's activation threshold and must be matched to the expected intracellular concentrations of the target metabolite.
- Operating Range (Detection Range) describes the span of ligand concentrations over which the biosensor exhibits a quantifiable response, bounded by lower and upper detection limits. The operating range must encompass the physiologically relevant concentrations of the target metabolite.
- Cooperativity (Hill Coefficient, n) describes the steepness of the dose-response curve. A Hill coefficient greater than 1 indicates positive cooperativity, which can create a sharper, more switch-like response desirable for certain dynamic regulation schemes.
- Signal-to-Noise Ratio measures the ratio between the output signal in the induced state and the variability of the signal in the uninduced state (basal expression). A high ratio is essential for reducing false positives.
- Response Time is the speed at which the output signal reaches its half-maximal value after induction. A faster response time is critical for dynamic regulation systems that need to react quickly to metabolic changes.
Table 1: Key Performance Parameters for Dose-Response Characterization
| Parameter | Description | Impact on Dynamic Regulation |
|---|---|---|
| Dynamic Range | Fold difference between basal and maximal output signal. | Determines the strength of the feedback control signal; a larger range offers a more powerful "on/off" switch. |
| Sensitivity (ECâ‚…â‚€/Kâ‚) | Ligand concentration for half-maximal response. | Must be tuned to match the metabolic precursor's physiological concentration to ensure activation at the correct flux level. |
| Operating Range | Concentration window of functional biosensor response. | Defines the metabolic "sweet spot" the biosensor can monitor and regulate. |
| Cooperativity (Hill n) | Steepness of the response curve. | A high Hill coefficient creates a sharper, more switch-like response, reducing intermediate states. |
| Basal Expression (Leakiness) | Output signal level in the absence of ligand. | High basal expression can waste cellular resources and cause metabolic imbalance before induction. |
| Response Time | Time to reach half-maximal output after induction. | Slower response can lead to oscillations and instability in a dynamic control circuit. |
Experimental Workflow for Dose-Response Characterization
A robust, standardized workflow is essential for generating reliable and reproducible dose-response data. The following protocol outlines the key steps from strain preparation to data analysis.
Strain Preparation and Cultivation
Begin by transforming the host strain (e.g., E. coli MG1655 or a production chassis) with the plasmid harboring the biosensor circuit. Key design elements typically include a constitutive promoter driving TF expression, the TF-responsive promoter controlling a reporter gene (e.g., GFP), and appropriate antibiotic resistance [58]. A single colony is used to inoculate a pre-culture in a defined medium, which is grown to saturation. The main experimental culture is then inoculated from the pre-culture at a standard dilution (e.g., 1:100) and grown to mid-exponential phase (OD₆₀₀ ≈ 0.5-0.6) [58].
Induction and Measurement
The culture is divided into multiple aliquots, each supplemented with a different, precisely defined concentration of the ligand. The ligand concentration series should span several orders of magnitude (e.g., from 0.001 mM to 100 mM) to adequately capture the lower limit, dynamic range, and saturation point of the biosensor [36] [58]. Following induction, the cultures are incubated for a defined period to allow for full reporter protein expression and maturation. This duration must be determined empirically and kept constant across all samples.
Data Collection and Analysis
For each induced culture, measure both the optical density (OD₆₀₀) and the reporter signal (e.g., fluorescence for GFP). The output is calculated as the reporter signal normalized by the OD. To generate the dose-response curve, plot the normalized output (y-axis) against the ligand concentration (x-axis) on a logarithmic scale. The resulting data can be fitted to a four-parameter Hill equation model to extract the key performance parameters.
Figure 1: Experimental workflow for characterizing biosensor dose-response curves, from strain preparation to quantitative analysis.
Tuning Strategies for Optimal Performance in Different Hosts
A biosensor characterized in a model host often requires tuning to function optimally in a production-relevant chassis. Performance can be fine-tuned through rational engineering of the biosensor's genetic components.
Table 2: Strategies for Tuning Biosensor Performance
| Component | Tuning Method | Primary Parameter(s) Affected | Mechanism of Action |
|---|---|---|---|
| Promoter | Modify operator sequence, number, or location; mutate -35/-10 regions. | Sensitivity, Dynamic Range, Cooperativity [6] | Alters the binding affinity of the TF and the recruitment efficiency of RNA polymerase. |
| Transcription Factor (TF) | Directed evolution or rational design of the ligand-binding domain (LBD). | Specificity, Sensitivity [6] [36] | Changes the affinity and selectivity of the TF for its ligand. |
| Ribosome Binding Site (RBS) | Engineer the RBS upstream of the TF or reporter gene. | Dynamic Range, Basal Expression [6] | Modulates the translation rate, thereby controlling the intracellular concentration of the TF or reporter. |
| Plasmid | Vary plasmid copy number. | Sensitivity, Signal Intensity [3] | Alters the gene dosage of both the TF and the reporter, impacting their expression levels. |
Promoter Engineering: The TF-responsive promoter is a primary tunable element. Changing the sequence, number, or position of TF operator sites within the promoter can significantly alter TF-binding affinity and thus the dose-response relationship [6] [2]. For instance, introducing point mutations in the operator site can reduce TF affinity, right-shifting the curve and increasing the operational range. Similarly, engineering the core -35 and -10 promoter regions can modulate the basal transcription rate and output intensity.
Transcription Factor Engineering: The TF itself can be engineered to adjust its affinity for the ligand (sensitivity) or its DNA operator (dynamic range). Directed evolution is a powerful strategy, involving creating mutant TF libraries and applying high-throughput screening to select variants with desired properties [36] [2]. Rational, computer-aided design is also emerging; for example, alanine scanning can identify key residues in the ligand-binding domain, which can then be mutated to alter specificity and sensitivity [36].
Translational Control (RBS Engineering): The translation rate of the TF protein is a critical factor. A TF expressed at very low levels may not effectively regulate its promoter, while overexpression can lead to high basal expression (for repressors) or constitutive activation (for activators) [6]. Engineering the RBS that controls TF translation allows for fine-tuning of TF abundance, optimizing the dynamic range and reducing leakiness.
Figure 2: Modular tuning strategies for optimizing key biosensor performance parameters.
Successful development and characterization of TF-based biosensors rely on a suite of key reagents, computational tools, and genetic resources.
Table 3: Research Reagent Solutions for Biosensor Characterization
| Category | Item | Function/Application |
|---|---|---|
| Genetic Parts | Constitutive Promoters (J23100, Pcons), RBS libraries, Fluorescent Reporters (GFP, mCherry), Terminators | Modular components for assembling and tuning the biosensor circuit. |
| Host Strains | E. coli MG1655 (standard), E. coli BL21 (protein expression), E. coli DH10B (cloning), Specialized production chassis (e.g., C. glutamicum) | Relevant chassis for testing biosensor performance and orthogonality. |
| Cloning Systems | CRISPR-Cas9 kits, Seamless cloning kits (e.g., Gibson Assembly), Site-directed mutagenesis kits | For precise construction and genome integration of biosensor circuits. |
| Analytical Tools | Microplate Reader (fluorescence, absorbance), Flow Cytometer, LC-MS/MS | Quantifying reporter output and validating intracellular metabolite concentrations. |
| Databases | RegulonDB, PRODORIC, P2TF, SM-TF | Source of well-characterized TFs, operator sequences, and regulon information [10] [9]. |
| Software/Tools | Cello (genetic circuit design), DeepTFactor (TF prediction), AlphaFold2 (protein structure prediction) | Computational design and in silico prediction for biosensor optimization [10] [8]. |
Case Study: Tuning a Lysine Biosensor for Dynamic Regulation
A 2025 study on a lysine biosensor for cadaverine production in E. coli provides an excellent example of this characterization and tuning pipeline in action [58]. The initial biosensor, based on the native CadC transcriptional activator and its promoter Pcad, exhibited a low dynamic range and was only functional under acidic conditions (pH < 5.8), making it unsuitable for standard fermentation.
To overcome these limitations, a multi-level optimization strategy was employed:
- TF Engineering: Key point mutations (e.g., Y47W/R89A) were introduced into the CadC protein based on its structural model. This expanded the biosensor's dynamic range by 3.3-fold and extended its functional pH range.
- Promoter Engineering: The native Pcad promoter was modified to fine-tune the interaction with the engineered TF, further improving the response to lysine.
- Characterization: The performance of the optimized biosensor variant was thoroughly characterized, showing a significantly widened operating range from 10â»â´ mM to 10 mM lysine.
When this tuned biosensor was implemented to dynamically regulate a cadaverine biosynthesis pathway, it resulted in a 48.1% increase in product titer (33.19 g/L) and a 21.2% improvement in cell growth compared to a constitutively expressed control [58]. This case underscores the critical impact of thorough dose-response characterization and strategic tuning on achieving successful dynamic regulation in bioproduction.
The precision of dynamic regulation in metabolic engineering is directly contingent on a deep and quantitative understanding of TF-based biosensor performance. By systematically characterizing the dose-response relationship and applying rational tuning strategies—such as promoter engineering, TF evolution, and RBS optimization—researchers can transform a poorly functioning genetic part into a robust, host-optimized biosensor. The experimental frameworks, parameters, and toolkits outlined in this guide provide a roadmap for this essential process. As the field advances, the integration of computational design and machine learning with high-throughput characterization will further accelerate the development of precision biosensors, paving the way for more efficient and intelligent microbial cell factories.
Biosensors are indispensable tools in synthetic biology, metabolic engineering, and therapeutic development, serving as critical interfaces for detecting cellular metabolites and regulating biological processes. For researchers focused on dynamic regulation, selecting the appropriate biosensor technology is paramount to achieving precise control over metabolic pathways and cellular functions. This whitepaper provides a comprehensive technical comparison of three prominent biosensor classes—transcription factor (TF)-based, aptamer-based, and FRET-based systems—framed within the context of dynamic regulation research. We examine their fundamental operating principles, performance characteristics, implementation requirements, and specific applicability to drug development and microbial engineering, supported by experimental protocols and quantitative performance data to guide selection for specific research applications.
Fundamental Principles and Mechanisms
Transcription Factor-Based Biosensors
TF-based biosensors are genetically encoded systems that utilize allosteric transcription factors to detect intracellular metabolites and regulate gene expression accordingly. Their mechanism involves three core steps: (1) analyte recognition through specific binding to the TF, (2) signal transduction via conformational changes that alter DNA-binding affinity, and (3) output generation through modulation of reporter gene expression [1] [8]. These systems naturally integrate with cellular regulatory networks, making them particularly valuable for dynamic metabolic engineering applications where real-time adjustment of pathway fluxes is required [59] [10].
The operational logic of TF-based biosensors depends on their inherent design architecture, which can follow multiple modes including repression of activator TF, activation of repressor TF, repression of repressor TF, or activation of activator TF [10]. This versatility enables their implementation in diverse genetic circuit configurations for sophisticated control schemes. For instance, in microbial cell factories, TF-based biosensors can dynamically balance metabolic fluxes by fine-tuning the expression of pathway enzymes without impeding cell growth, significantly enhancing the production of valuable compounds like aromatic amino acids and their derivatives [59].
Aptamer-Based Biosensors
Aptamer-based biosensors utilize synthetic single-stranded DNA or RNA oligonucleotides as recognition elements that undergo structural reorganization upon binding to specific targets [60] [61]. These systems leverage the Systematic Evolution of Ligands by Exponential Enrichment (SELEX) process to generate high-affinity binding molecules with antibody-comparable specificity [60]. Unlike protein-based systems, aptamers offer superior chemical stability, ease of synthesis and modification, and reduced batch-to-batch variability, making them attractive for diagnostic and environmental monitoring applications [62] [61].
The signaling mechanism in aptasensors typically involves binding-induced conformational changes that alter the spatial relationship between reporter molecules, such as fluorophores and quenchers, or affect electron transfer efficiency in electrochemical detection platforms [60]. Advanced SELEX variants including capture-SELEX, capillary electrophoresis-SELEX (CE-SELEX), and nitrocellulose SELEX have expanded the range of detectable targets, particularly for small molecules that lack functional groups for immobilization [60]. These developments have enabled the creation of robust sensing platforms for diverse targets including heavy metals, antibiotics, foodborne pathogens, and protein biomarkers associated with neurodegenerative diseases [62] [60].
FRET-Based Biosensors
Förster Resonance Energy Transfer (FRET)-based biosensors operate through distance-dependent non-radiative energy transfer between a donor fluorophore and an acceptor molecule when separated by 1-10 nm [63] [64]. FRET efficiency exhibits an inverse sixth-power relationship with distance, making these sensors exquisitely sensitive to molecular-scale distance changes resulting from binding events, enzymatic activities, or conformational shifts [63]. This physical principle enables real-time monitoring of biological processes including protein-protein interactions, DNA hybridization, and enzymatic activities with high spatial and temporal resolution [63].
FRET-based systems typically employ paired fluorophores such as organic dyes (FAM/TAMRA, Cy3/Cy5), fluorescent proteins (CFP/YFP), or nanomaterials (quantum dots, gold nanoparticles) [63] [64]. Their implementation spans diverse applications from intracellular signaling studies to diagnostic platforms, particularly in cancer biomarker detection where they facilitate sensitive exosome analysis in liquid biopsies [64]. A significant advantage of FRET biosensors is their ability to detect molecular interactions without requiring sample manipulation or separation steps, enabling homogeneous assay formats and real-time monitoring of biological processes in living cells [63].
Performance Comparison and Technical Specifications
Table 1: Comparative Performance Metrics of Biosensor Technologies
| Parameter | TF-Based Biosensors | Aptamer-Based Biosensors | FRET-Based Biosensors |
|---|---|---|---|
| Detection Limit | Varies by TF; ~nM-μM range for metabolites [59] | Excellent; pM-nM range for various targets [60] | Superior; pM-nM range for exosomes, ions [63] [64] |
| Dynamic Range | 2-100 fold; tunable via promoter/operator engineering [1] [3] | >100 fold; highly tunable [60] | Limited by fluorophore properties; moderate [63] |
| Response Time | Minutes to hours (requires transcription/translation) [3] | Seconds to minutes (conformational change) [60] | Microseconds to seconds (instant fluorescence response) [63] |
| Target Scope | Natural intracellular metabolites, some engineered for new targets [10] | Extremely broad: ions, small molecules, proteins, cells [60] [61] | Molecular interactions, conformational changes, proximity assays [63] [64] |
| Specificity | Moderate; can exhibit cross-reactivity [1] | Excellent; engineered for minimal cross-reactivity [60] | High; depends on recognition element specificity [63] |
| Stability | Good in cellular environments; subject to proteolysis [1] | Excellent; thermal and chemical stability [60] [61] | Variable; photobleaching can limit long-term use [63] |
Table 2: Application Suitability Across Research Domains
| Application Domain | TF-Based Biosensors | Aptamer-Based Biosensors | FRET-Based Biosensors |
|---|---|---|---|
| Dynamic Metabolic Regulation | Excellent; inherent genetic integration [59] [3] | Limited; primarily sensing function | Moderate; can monitor but not regulate |
| High-Throughput Screening | Good; fluorescent/colorimetric outputs [1] | Excellent; various signal outputs [60] | Excellent; high sensitivity [63] |
| In Vivo/Intracellular Monitoring | Excellent; genetically encoded [10] | Moderate; delivery challenges | Good; requires fluorophore introduction [64] |
| Diagnostic Applications | Limited | Excellent; various platforms [62] [60] | Excellent; especially clinical biomarkers [64] |
| Environmental Monitoring | Limited | Excellent; robust detection [61] | Moderate; matrix effects [63] |
| Therapeutic Development | Promising for engineered cell therapies [3] | Good for biomarker detection [62] | Excellent for drug screening [63] |
Experimental Implementation Guide
Protocol for TF-Based Biosensor Construction and Validation
Objective: Implement a TF-based biosensor for dynamic regulation of metabolic pathways in E. coli, specifically targeting aromatic compound production [59].
Materials:
- Bacterial strains (e.g., E. coli MG1655 or derivative)
- Plasmid vectors with modular cloning sites
- TF genes and promoter sequences (e.g., TrpR, HucR, PadR)
- Reporter genes (e.g., GFP, RFP, antibiotic resistance)
- Target metabolites for validation
- Fluorescence measurement equipment
Procedure:
Biosensor Assembly:
- Clone the TF gene under a constitutive promoter into your expression vector
- Insert the corresponding TF-regulated promoter upstream of your reporter gene
- Include the metabolic pathway genes under control of the TF-regulated promoter for dynamic regulation applications
Characterization:
- Transform the constructed plasmid into your production strain
- Culture cells and expose to varying concentrations of target metabolite
- Measure reporter output (fluorescence, absorbance) at regular intervals
- Generate a dose-response curve to determine dynamic range and EC50
Dynamic Regulation Validation:
- Cultivate engineered strains in production medium
- Monitor metabolite accumulation and pathway gene expression over time
- Compare production yields against constitutively expressed controls
Troubleshooting Tips:
- High background signal may require promoter engineering to reduce leakiness
- Limited dynamic range can be addressed by RBS optimization or TF engineering
- Cross-talk with native regulation may necessitate chassis engineering
Protocol for Aptamer-Based Biosensor Development
Objective: Develop an aptamer-based electrochemical biosensor for detection of food contaminants or disease biomarkers [60].
Materials:
- Synthetic aptamer sequences (from SELEX selection)
- Functionalized electrodes (gold, carbon, or screen-printed)
- Electrochemical cell and potentiostat
- Target analytes and potential interferents
- Nanomaterials for signal enhancement (e.g., AuNPs, graphene oxide)
Procedure:
Aptamer Immobilization:
- Thiol- or amino-modified aptamers are chemisorbed onto electrode surfaces
- Block nonspecific binding sites with BSA or similar blocking agents
- Validate surface coverage through electrochemical impedance spectroscopy
Assay Optimization:
- Incubate functionalized electrodes with varying target concentrations
- Optimize binding time, temperature, and buffer conditions
- Measure signal changes (current, impedance, potential)
Sensor Validation:
- Test sensor specificity against structurally similar compounds
- Determine detection limit and linear range in buffer and complex matrices
- Assess sensor reproducibility and stability over time
Advanced Applications:
- For in vivo monitoring, conjugate aptamers with signal-generating nanomaterials
- Implement in microfluidic devices for point-of-care testing
- Develop multiplexed platforms using different aptamers on arrayed electrodes
Protocol for FRET-Based Biosensor Implementation
Objective: Construct a FRET-based biosensor for detecting exosomal biomarkers for cancer diagnostics [64].
Materials:
- Donor and acceptor fluorophore pairs (e.g., FAM/BHQ1, Cy3/Cy5)
- Recognition elements (antibodies, aptamers) for target exosomal markers
- Purified exosome samples or clinical specimens
- Fluorescence spectrometer or microscope with FRET capability
- Surface modification reagents for nanoparticle-based FRET
Procedure:
Biosensor Design:
- Conjugate donor fluorophore to primary recognition element
- Position acceptor fluorophore within FRET distance (1-10 nm)
- For exosome detection, use antibodies against CD63, CD81, or specific tumor markers
Assay Configuration:
- Incubate FRET pair with target exosomes in solution or on surfaces
- Allow binding-induced distance changes to alter FRET efficiency
- Measure donor quenching and/or acceptor sensitization
Signal Detection:
- Use fluorescence spectroscopy with appropriate filter sets
- Calculate FRET efficiency using acceptor photobleaching or ratio-metric methods
- Generate calibration curves with standard exosome preparations
Advanced Implementation:
- Utilize quantum dots or upconversion nanoparticles as donors for enhanced photostability
- Implement in microfluidic devices for automated processing
- Combine with smartphone-based detection for point-of-care applications
Research Reagent Solutions
Table 3: Essential Research Reagents for Biosensor Development
| Reagent Category | Specific Examples | Research Application | Key Suppliers/References |
|---|---|---|---|
| Transcription Factors | TrpR (L-tryptophan), HucR (vanillin), PadR (p-Coumaric acid) [59] | Dynamic regulation of aromatic compound pathways | Academic labs; Addgene for genetic parts |
| Aptamer Sequences | Antibiotic-specific, heavy metal-binding, protein biomarker aptamers [60] | Molecular recognition in diagnostic and environmental sensors | Custom synthesis from IDT, Sigma; aptamer databases |
| Fluorophore Pairs | FAM/BHQ1, Cy3/Cy5, CFP/YFP, Quantum Dots/AuNPs [63] [64] | FRET-based proximity and interaction assays | Thermo Fisher, ATTO-TEC, Sigma-Aldrich |
| Reporter Genes | GFP, RFP, mCherry, luciferase, LacZ [59] [10] | Visualizing biosensor activation and output signals | Addgene, ATCC, commercial plasmid collections |
| Nanomaterials | Gold nanoparticles, graphene oxide, MOFs, UCNPs [60] [64] | Signal amplification and platform functionalization | Sigma-Aldrich, NanoComposix, ACS Materials |
| Engineering Tools | Directed evolution kits, modular cloning systems, computational design software [1] [10] | Optimizing biosensor performance and dynamic range | NEB, Takara Bio, Cello computational tool |
Integration in Dynamic Regulation Research
For researchers focused on dynamic regulation, TF-based biosensors offer unique advantages due to their innate biological compatibility and capacity for closed-loop control of metabolic pathways [59] [3]. These systems enable autonomous optimization of flux distribution by responding to intracellular metabolite levels and adjusting enzyme expression accordingly, overcoming the limitations of static optimization approaches [59]. Notable implementations include the regulation of aromatic amino acid biosynthesis in E. coli, where TF-based biosensors have balanced precursor availability and pathway enzyme expression to enhance production of compounds like L-tryptophan, L-tyrosine, and valuable derivatives [59].
The integration of computational tools has significantly advanced TF-based biosensor applications in dynamic regulation. Platforms like Cello enable in silico design and optimization of genetic circuits, allowing researchers to simulate biosensor performance before experimental implementation [1] [8]. Additionally, machine learning approaches are being employed to predict TF specificity and design novel biosensors for metabolites lacking natural biosensors [10]. These computational strategies are particularly valuable for optimizing biosensor dynamic range, specificity, and orthogonality in complex regulatory networks [1] [10].
While aptamer and FRET-based systems excel in detection sensitivity and speed, their application in dynamic regulation is primarily limited to sensing functions rather than direct genetic control. However, these technologies can be integrated with TF-based systems to create hybrid platforms that leverage the strengths of each approach. For instance, aptamer-based sensors can detect extracellular metabolites that cannot be directly sensed by TFs, while FRET-based reporters can provide high-temporal resolution monitoring of pathway dynamics to inform genetic controller design [63] [60].
The selection of an appropriate biosensor platform depends fundamentally on the specific research requirements, particularly the intended application in sensing versus regulatory control. TF-based biosensors offer unparalleled capabilities for dynamic metabolic regulation in living cells, making them ideal for metabolic engineering and synthetic biology applications where autonomous control of pathway fluxes is desired. Aptamer-based systems provide exceptional versatility and sensitivity for diagnostic and environmental monitoring applications, with superior stability and customization potential. FRET-based biosensors deliver unmatched spatial and temporal resolution for studying molecular interactions and conformational changes, making them invaluable for mechanistic studies and clinical diagnostics.
For researchers focused on dynamic regulation, TF-based biosensors represent the most direct and biologically integrated approach, particularly when combined with computational design tools and directed evolution strategies to enhance their performance. Future developments in biosensor technology will likely focus on creating hybrid systems that combine the sensing strengths of aptamer and FRET-based approaches with the regulatory capabilities of TF-based systems, enabling more sophisticated control schemes for therapeutic and bioproduction applications.
Transcription factor (TF)-based biosensors are indispensable tools in synthetic biology and metabolic engineering, enabling dynamic regulation of genetic circuits, real-time monitoring of intracellular metabolites, and high-throughput screening of production strains [9] [8]. These biological devices function by combining a sensing component (typically an allosteric transcription factor or aTF) that detects a specific chemical input with a reporter module that produces a measurable output signal [9] [65]. The validation of these biosensors—ensuring their specificity, sensitivity, and dynamic range—is a critical step in their development cycle. This process heavily relies on the strategic use of public databases and computational resources to guide experimental design, predict performance, and interpret results. Within the broader context of dynamic regulation research, robust validation protocols ensure that biosensors can reliably control metabolic fluxes in microbial cell factories, thereby optimizing bioproduction processes [9] [15]. This technical guide provides researchers with a comprehensive framework for leveraging publicly available data and tools to validate TF-based biosensors effectively.
A critical first step in biosensor validation involves gathering existing knowledge on transcription factors, their effector molecules, and their DNA-binding specificities. The table below summarizes key public databases and their utility in the biosensor validation workflow.
Table 1: Essential Public Databases for Biosensor Research and Validation
| Database Name | Primary Focus | Key Data Types | Utility in Biosensor Validation |
|---|---|---|---|
| RegulonDB [9] | Transcriptional regulation in E. coli K-12 | TF binding sites, regulated genes, operons | Reference for validating TF-promoter interactions in a model organism |
| PRODORIC [9] | Prokaryotic gene regulation | Manually curated TF binding sites | Curated data for designing control experiments and verifying specificity |
| JASPAR [9] | TF binding profiles across species | Position Frequency Matrices (PFMs) | Predicting and validating DNA binding motifs for novel TFs |
| Bionemo [9] [66] | Biodegradation metabolism | Proteins/genes in biodegradation | Identifying TFs responsive to environmental pollutants or metabolic intermediates |
| RegPrecise [9] [66] | Prokaryotic regulons | Reconstructed regulons, TF binding sites | Comparative genomics to infer and validate regulatory interactions |
| SM-TF [9] | Structural data on TF complexes | 3D structures of small molecule-TF complexes | Informing mutational studies and engineering efforts via structural insights |
| AnimalTFDB / PlantTFDB [9] | TF classification in animals/plants | TF inventory, classification | Resource for researchers working with eukaryotic host systems |
These databases collectively help establish a "detectable input space," defining the known set of molecules that can be detected by TF-based biosensors [9]. Validation often begins by querying these resources to confirm the known properties of a biosensor's components before functional testing. Furthermore, databases like Sigmol (for Quorum Sensing molecules) and GroovDB provide specialized, curated information that can be used to source well-characterized genetic parts for biosensor construction, providing a positive control for validation experiments [9] [66].
Validation Methodologies and Workflows
Computational Validation and Prediction
Before embarking on wet-lab experiments, computational tools can predict biosensor function and guide design, making subsequent validation more efficient.
Table 2: Computational Tools for Biosensor Design and Preliminary Validation
| Tool Name | Methodology | Primary Function | Validation Application |
|---|---|---|---|
| Snowprint [38] | Bioinformatic algorithm based on inverted repeat conservation | Predicts operator sequences for transcriptional regulators | Identifies putative DNA binding sites for novel TFs, enabling promoter design for experimental testing |
| Sensbio [66] | Molecular similarity (Tanimoto score) & sequence similarity (BLAST) | Suggests potential TF-ligand pairs based on similarity to known pairs | Proposes putative effector molecules for orphan TFs and alternative inducers for known TFs |
| CiiDER [9] | Bioinformatics analysis | Predicts and analyzes transcription factor binding sites | In silico validation of designed promoter sequences |
| BART [9] | Bioinformatics tool | Predicts functional transcriptional regulators | Helps identify potential TFs that could regulate a gene of interest |
| DeepTFactor [9] | Deep Learning | Predicts sequence-derived transcription factors | Annotates and identifies putative TFs in genomic sequences |
Snowprint exemplifies a powerful validation tool. Its workflow begins with a regulator's protein accession ID, fetches its genetic context, and extracts the inter-operon region upstream of the regulator. It then identifies a "seed operator" by searching for conserved inverted repeats and compares this seed against homologs to generate a consensus predicted operator with a conservation score [38]. This prediction can be validated experimentally using techniques like Electromobility Shift Assays (EMSA) or DNase footprinting [38]. Benchmarking has shown that Snowprint produces predictions significantly similar to known operators (E-value < 0.01) for 58% of TetR-family regulators and 44% of MarR-family regulators [38].
Sensbio aids validation by leveraging chemical similarity. It uses molecular fingerprints and the Tanimoto similarity score to compare a query molecule against its database of known TF-ligand pairs [66]. A high score suggests that the query molecule may be detected by the same TF, which can be directly tested. This is particularly useful for identifying potential cross-reactivity or non-specific activation, a key aspect of validating biosensor specificity.
The following diagram illustrates a general workflow for discovering and validating a novel biosensor, integrating database mining, computational prediction, and experimental steps.
Experimental Validation and Performance Tuning
Once a biosensor is designed and constructed, its performance must be quantitatively assessed through controlled experiments. Key performance criteria include [15]:
- Dynamic Range: The fold-change between the "ON" and "OFF" output states.
- Sensitivity: The concentration of ligand required to elicit a half-maximal response.
- Specificity: The biosensor's ability to distinguish the target ligand from structurally similar molecules.
- Detection Range: The span of ligand concentrations over which the biosensor responds.
A common experimental protocol for validating a repressive biosensor in yeast (e.g., using the FapR/fapO system for malonyl-CoA) involves the following steps [67]:
- Strain Construction: Clone the TF (e.g., FapR) fused to a transcriptional activation domain (e.g., Med2) and integrate it into the host genome. The reporter gene (e.g., GFP) is placed under a promoter containing the TF's operator sites.
- Ligand Induction: Treat cultures with graduated concentrations of a ligand or an agent that modulates the intracellular concentration of the target metabolite. For malonyl-CoA biosensors, cerulenin is used to inhibit fatty acid synthesis, leading to malonyl-CoA accumulation [67].
- Output Measurement: Measure the fluorescence intensity of the cultures using a flow cytometer or microplate reader after a defined incubation period.
- Data Analysis: Calculate the repression ratio as
(Fluorescence_OFF - Fluorescence_ON) / Fluorescence_OFF. Plot the dose-response curve to determine the dynamic range, sensitivity, and detection range [67].
To optimize performance, systematic engineering is often required. For a repressive biosensor, this can involve [67]:
- Activator Domain Screening: Comparing different activation domains (e.g., Gal4, VP16, Med2) for their effect on the dynamic range.
- Promoter Engineering: Varying the number, position, and sequence of the operator sites within the promoter.
- TF Expression Tuning: Modifying the expression level of the TF itself to minimize leakiness and maximize response.
For altering ligand specificity, a computation-guided workflow can be highly effective, as demonstrated for engineering the BenM TF to respond to adipic acid instead of its native ligand, cis,cis-muconic acid [46]:
- Ligand Docking: Use molecular docking software to model the target ligand (adipic acid) into the TF's ligand-binding pocket.
- Identify Hotspot Residues: Identify amino acid residues in the binding pocket that, if mutated, would improve complementarity with the new ligand.
- Library Creation & Screening: Create a site-saturation mutagenesis library at the identified residue(s) and screen for clones with the desired ligand specificity.
- Validation & Analysis: Characterize the performance of hits and use molecular dynamics simulations to understand the structural basis for the altered specificity [46].
The Scientist's Toolkit: Research Reagent Solutions
The table below lists essential materials and reagents used in the featured biosensor validation experiments, along with their specific functions.
Table 3: Research Reagent Solutions for Biosensor Validation
| Reagent / Material | Function in Validation | Example Use Case |
|---|---|---|
| Cerulenin [67] | Inhibits fatty acid synthesis, leading to intracellular accumulation of malonyl-CoA. | Used as an indirect inducer to validate malonyl-CoA repressive biosensors in S. cerevisiae. |
| Gal4 Activation Domain (AD) [67] | A classic transcriptional activation domain fused to the DNA-binding TF to create a synthetic activator. | Part of the initial design for a malonyl-CoA repressive biosensor. |
| Med2 Activation Domain (AD) [67] | A subunit of the yeast mediator complex that acts as a highly efficient transcriptional activator. | Engineered into a FapR-based biosensor, resulting in a superior dynamic range compared to Gal4-AD. |
| VP64-p65-Rta (VPR) AD [67] | A synthetic, tripartite transcriptional activator known for high efficiency. | Fused to FapR and evaluated for its ability to create a high-performance repressive biosensor. |
| Green Fluorescent Protein (GFP) [67] [65] | A reporter module that produces a quantifiable fluorescent output signal. | Standard reporter for measuring biosensor activity via flow cytometry or fluorescence spectroscopy. |
| Flow Cytometer | Instrument for measuring fluorescence intensity of individual cells in a population. | Enables high-resolution, single-cell analysis of biosensor performance and population heterogeneity. |
The rigorous validation of transcription factor-based biosensors is a multi-faceted process that strategically integrates computational resources with experimental biology. Public databases provide the foundational knowledge of TF-ligand interactions and DNA binding specificities, while modern bioinformatic tools like Snowprint and Sensbio enable predictive design and in silico hypothesis testing. Experimental validation, guided by these resources, relies on quantitative assessments of dynamic range, sensitivity, and specificity, often requiring iterative optimization of genetic components. As the field advances, the integration of computational modeling, machine learning, and high-throughput screening will further streamline the biosensor validation pipeline, accelerating their application in dynamic metabolic regulation and the development of advanced microbial cell factories.
Transcription factor-based biosensors (TFBs) are indispensable tools in synthetic biology and metabolic engineering, serving as critical components for real-time monitoring, high-throughput screening, and dynamic regulation of metabolic pathways [14] [4]. These genetically encoded devices function by converting the intracellular concentration of a specific target metabolite into a measurable output signal, typically fluorescence or luminescence [9] [8]. The ability of TFBs to provide a direct, real-time link between the metabolic state of a cell and a quantifiable readout has positioned them as fundamental enabling technologies for the development of efficient microbial cell factories, particularly for the production of high-value compounds like advanced biofuels and natural products [14] [4].
However, the journey from a novel biosensor concept to a validated, high-performance genetic circuit is complex and requires systematic characterization. Wild-type biosensors rarely possess the requisite sensitivity, specificity, or dynamic range for immediate application in demanding industrial or research settings [9] [8]. This case study provides an in-depth technical guide for the comprehensive validation of a novel TFB for a target metabolite. Framed within the broader context of advancing dynamic regulation research, we detail a structured workflow encompassing computational design, experimental characterization of key performance parameters, and ultimate deployment in functional applications such as high-throughput screening and dynamic metabolic control. The protocols and analytical frameworks presented herein are designed to equip researchers with the methodologies necessary to transform a putative biosensor into a robust and reliable tool for metabolic engineering.
Biosensor Components and Design Principles
A functional transcription factor-based biosensor is a modular genetic circuit constructed from several core components that work in concert to transduce a chemical signal into gene expression [4] [8]. The sensing module consists of an allosteric transcription factor (TF) that specifically binds to a target metabolite (the inducer or ligand). This binding event triggers a conformational change in the TF. The genetic module includes a promoter region containing a specific DNA sequence, the transcription factor binding site (TFBS), to which the TF binds. Depending on the TF's nature, ligand binding can either activate or repress its ability to regulate transcription. Finally, the output module is a reporter gene positioned downstream of this promoter, whose expression is controlled by the TF-ligand interaction, producing a detectable signal such as green fluorescent protein (GFP) for fluorescence or luciferase for luminescence [9] [4].
The design phase is critical for establishing a successful biosensor. Table 1 summarizes the essential genetic parts and their functions. Prior to experimental work, in silico analysis is highly recommended. Databases such as RegulonDB for E. coli, JASPAR for TF binding profiles, and BioNemo for biodegradation metabolism proteins can be mined to identify candidate TFs and their cognate promoters and binding sites [9]. Furthermore, tools like DeepTFactor, which uses deep learning to predict whether a protein functions as a transcription factor, can guide the identification of novel sensing elements from genomic or metagenomic data [9]. For non-model metabolites where a native TF may not exist, emerging strategies include the construction of chimeric biosensors through domain swapping or the application of de novo protein design to create entirely novel ligand-binding domains [49].
Table 1: Key Genetic Components of a Transcription Factor-Based Biosensor
| Component | Description | Function | Design Considerations |
|---|---|---|---|
| Transcription Factor (TF) | Allosteric protein (e.g., from TetR, LacI, AraC families). | Senses the target metabolite and undergoes conformational change. | Ligand specificity, sensitivity, and orthogonality to the host chassis [9] [8]. |
| Promoter & TF Binding Site (TFBS) | Specific DNA sequence upstream of the reporter gene. | Site for TF binding, controlling the initiation of transcription. | Strength of the promoter and affinity of the TF for the TFBS influence the dynamic range [4]. |
| Reporter Gene | Gene encoding a detectable protein (e.g., GFP, RFP, Luciferase). | Generates a quantifiable output signal correlated with ligand concentration. | Signal intensity, maturation time, and suitability for high-throughput detection [4] [8]. |
| Ribosome Binding Site (RBS) | Sequence upstream of the reporter gene. | Controls the translation initiation rate of the reporter. | RBS strength can be tuned to optimize translation efficiency and output signal [4]. |
The logical relationship and workflow for designing and validating a biosensor are complex. The following diagram illustrates the key stages from initial design to final application, highlighting the iterative "Design-Build-Test-Learn" cycle central to synthetic biology.
Performance Characterization and Key Metrics
Once a biosensor circuit is constructed, its performance must be rigorously quantified to determine its suitability for application. Characterization involves measuring the biosensor's output across a gradient of inducer concentrations to generate an input-output response curve, which is typically fitted to a Hill equation model [9] [4]. From this curve, several critical quantitative parameters are derived, as detailed in Table 2.
Table 2: Key Quantitative Performance Metrics for Biosensor Validation
| Performance Metric | Definition | Experimental Measurement | Ideal Target |
|---|---|---|---|
| Dynamic Range | The fold-change between the maximum and minimum output signal [4]. | (Outputmax - Outputmin) / Output_min | A large fold-change (e.g., >50) for easy distinction [8]. |
| Sensitivity (ECâ‚…â‚€/KD) | The ligand concentration required to generate a half-maximal response (ECâ‚…â‚€), related to the TF's binding affinity (KD) for the ligand [4]. | Ligand concentration at 50% of the dynamic range on the dose-response curve. | Matched to the expected intracellular metabolite concentration. |
| Operational Range | The range of ligand concentrations over which the biosensor response is dynamic and measurable [4]. | Concentration range between ~10% and ~90% of the maximum output. | Broad enough to cover physiological relevant concentrations. |
| Specificity | The ability to respond exclusively to the target metabolite and not to structurally similar analogs [4]. | Measure response to the target vs. a panel of potential cross-reactants. | High specificity to avoid false-positive signals in complex media. |
| Orthogonality | The lack of interference with the host's native regulatory networks [9]. | Assess host cell growth and native gene expression with biosensor present. | High orthogonality for predictable performance in the host chassis. |
A well-characterized biosensor will exhibit a sigmoidal dose-response curve. The point of half-maximal response is the ECâ‚…â‚€, a key measure of sensitivity. The operational range spans the concentrations where the response is most dynamic, and the maximum output level determines the dynamic range. Specificity is validated by challenging the biosensor with other compounds; a specific biosensor will only show significant response to its intended target.
Experimental Protocols for Biosensor Validation
This section provides detailed methodologies for the key experiments required to populate the performance metrics outlined in Table 2.
Protocol: Generating a Dose-Response Curve
This protocol is fundamental for determining the dynamic range, sensitivity, and operational range of the biosensor [4].
- Strain and Culture Preparation: Transform the biosensor plasmid into the appropriate microbial host (e.g., E. coli). Inoculate a single colony into liquid culture medium and grow overnight.
- Inducer Dilution Series: Prepare a serial dilution of the target metabolite (inducer) in culture medium across a broad concentration range (e.g., from 0 μM to 100 mM), spanning several orders of magnitude. Include a negative control with no inducer.
- Cultivation and Induction: Dilute the overnight culture to a standard optical density (OD) and aliquot it into the inducer solutions. Grow the cultures under defined conditions (temperature, shaking).
- Signal Measurement: Once the cultures reach the mid-exponential or stationary phase, measure both the cell density (OD) and the reporter signal (e.g., fluorescence for GFP, luminescence for luciferase) for each culture.
- Data Normalization: Normalize the reporter signal to the cell density to account for growth differences (e.g., Fluorescence/OD).
- Curve Fitting: Plot the normalized output signal against the inducer concentration on a logarithmic scale. Fit the data to the Hill equation to derive the key parameters: maximum output, minimum output, Hill coefficient (n), and ECâ‚…â‚€.
Protocol: Specificity and Cross-Reactivity Testing
This assay evaluates the biosensor's specificity [4].
- Compound Selection: Compile a panel of compounds including the target metabolite, its known structural analogs, pathway precursors, and final products.
- Culture and Induction: Follow the same procedure as in Protocol 4.1, but instead of a dilution series, use a single, relevant concentration (e.g., the ECâ‚…â‚€) for each compound in the panel.
- Signal Measurement and Analysis: Measure the normalized output signal for each compound. The response to any non-target compound should be significantly lower (e.g., <20%) than the response to the target metabolite at the same concentration.
Protocol: Assessing Orthogonality and Host Impact
This protocol ensures the biosensor functions without detrimental effects on the host [9].
- Strain Comparison: Compare the growth curve (OD over time) of the host strain harboring the biosensor plasmid to a control strain (with an empty plasmid or a non-functional version).
- Fit Analysis: Calculate the maximum growth rate and final biomass yield for both strains. A significant impairment in the biosensor strain indicates a high metabolic burden or unintended interference with native processes.
- Contextual Performance: For dynamic regulation applications, it is critical to test the biosensor's performance not just in a plug-and-play context, but within the full metabolic pathway. This involves evaluating its response to fluctuations in the pathway intermediate it senses, ensuring it can effectively trigger regulation under realistic production conditions [68].
Application in Metabolic Engineering
Validated high-performance biosensors are deployed in two primary applications that accelerate metabolic engineering.
High-Throughput Screening
TFBs enable the rapid screening of vast genomic or mutant libraries to identify production strains. The biosensor is engineered to link the production of the target metabolite to a survival gene (e.g., antibiotic resistance) or an easily sortable marker like GFP [4] [8]. When a library of pathway variants is created, cells with high metabolic flux will produce more of the target, triggering a stronger biosensor response. This allows researchers to use fluorescence-activated cell sorting (FACS) to physically isolate the top producers from a population of millions of cells in a matter of hours, dramatically accelerating the strain optimization process [14] [4].
Dynamic Pathway Regulation
Biosensors can be used to implement closed-loop, dynamic control systems within cell factories [4] [68]. In this configuration, the biosensor does not simply report via a reporter gene, but instead controls the expression of a key rate-limiting enzyme in the biosynthetic pathway. When the intracellular concentration of a critical intermediate becomes too high (indicating a bottleneck or imbalance), the biosensor activates to express the enzyme, alleviating the bottleneck. Conversely, when the concentration is low, the biosensor turns off, preventing wasteful resource allocation. This dynamic feedback control provides robustness to the production system, automatically compensating for metabolic fluctuations and perturbations, and ultimately leading to higher titers and yields [4] [68]. The workflow for applying a validated biosensor is a logical progression from screening to sophisticated regulation.
The Scientist's Toolkit: Research Reagent Solutions
The development and application of transcription factor-based biosensors rely on a suite of essential reagents and tools. The following table details key materials and their functions in a typical biosensor research workflow.
Table 3: Essential Research Reagents and Tools for Biosensor Development
| Reagent / Tool | Function in Biosensor Workflow | Examples / Notes |
|---|---|---|
| Allosteric Transcription Factors | Core sensing element; binds the target metabolite. | TetR, AraC, LysR family TFs; sourced from databases like RegulonDB or identified via homology mining [9]. |
| Reporter Genes | Generates measurable output for quantification. | GFP, RFP (fluorescence), Luciferase (luminescence), LacZ (colorimetry) [4] [8]. |
| Expression Vectors & Plasmids | Genetic backbone for assembling and hosting the biosensor circuit. | Standard (e.g., pBR322 origin) or high-copy number plasmids; choice affects gene dosage and biosensor performance [4]. |
| Model Host Organisms | Chassis for biosensor expression and functional testing. | Escherichia coli (common), Saccharomyces cerevisiae (yeast), other non-model production hosts [14] [9]. |
| Metabolite Standards | Used for calibration and generating dose-response curves. | High-purity target metabolite and structural analogs for specificity testing. |
| Microplate Readers | Instrumentation for high-throughput measurement of output signals (fluorescence, luminescence, OD). | Essential for efficiently collecting dose-response data and screening assays. |
| Flow Cytometers (FACS) | Enables high-throughput screening and isolation of high-producing cells from large libraries. | Critical for applying biosensors in directed evolution and library screening [4]. |
Conclusion
Transcription factor-based biosensors represent a rapidly advancing frontier in synthetic biology, offering unprecedented capability for the dynamic regulation of cellular processes. The synthesis of knowledge across the four intents confirms that these devices are transitioning from simple proof-of-concept tools to robust, engineerable systems capable of addressing complex challenges in biomanufacturing and biomedicine. Key takeaways include the critical importance of fine-tuning biosensor components to achieve desired performance, the growing role of computational tools and AI in accelerating design and optimization, and the successful application of these biosensors in creating more efficient microbial cell factories. Future progress hinges on expanding the library of characterized transcription factors through multi-omics mining and de novo design, enhancing orthogonality to prevent host interference, and integrating biosensors with AI for real-time, closed-loop control of metabolic states. These advancements promise to unlock new paradigms in smart therapeutics, sustainable chemical production, and sophisticated diagnostic platforms, firmly establishing TF-based biosensors as indispensable tools in the bioeconomy.
References
- 1. Microbial Transcription Factor-Based Biosensors: Innovations ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12024804/]
- 2. Advances in engineering and optimization of transcription ... [https://www.sciencedirect.com/science/article/abs/pii/S0958166922000878]
- 3. Challenges and Opportunities in Smart Biosensing for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12455640/]
- 4. A molecular-guided approach for advanced biofuel synthesis [https://www.sciencedirect.com/science/article/abs/pii/S0734975024000338]
- 5. Transcription factor-based biosensors enlightened by the ... [https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2015.00648/full]
- 6. Applications and Tuning Strategies for Transcription Factor ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10136082/]
- 7. Engineered TtgR-Based Whole-Cell Biosensors for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12384351/]
- 8. Innovations from Design to Applications in Synthetic Biology [https://www.mdpi.com/2079-6374/15/4/221]
- 9. Transcription factor-based biosensors for screening and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9939652/]
- 10. Transcription factor-based biosensors for screening and ... [https://www.frontiersin.org/journals/bioengineering-and-biotechnology/articles/10.3389/fbioe.2023.1118702/full]
- 11. Transcriptional gene expression: Activators and repressors [https://rotel.pressbooks.pub/genetics/chapter/transcriptional-gene-expression-activators-and-repressors/]
- 12. Emergence of activation or repression in transcriptional ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11160767/]
- 13. Transcription factor-based biosensors for detection of ... [https://www.sciencedirect.com/science/article/pii/S187167842300047X]
- 14. Advances in transcription factor-based biosensors for ... [https://pubmed.ncbi.nlm.nih.gov/41274426/]
- 15. Applications and Tuning Strategies for Transcription Factor ... [https://www.mdpi.com/2079-6374/13/4/428]
- 16. Highly multiplexed design of an allosteric transcription ... [https://www.nature.com/articles/s41467-024-54260-8]
- 17. At-home, Cell-free Synthetic Biology Education Modules for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11577157/]
- 18. Using Synthetic Biology to Make Cells Tomorrow's Test Tubes [https://pmc.ncbi.nlm.nih.gov/articles/PMC4837077/]
- 19. Synthetic Evolution of Metabolic Productivity Using ... [https://www.sciencedirect.com/science/article/abs/pii/S0167779916000354]
- 20. Principles of Genetic Circuit Design - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4230274/]
- 21. Construction of a genetic AND gate under a new standard for ... [https://bmcgenomics.biomedcentral.com/articles/10.1186/1471-2164-11-S4-S16]
- 22. Regulation of Protein Synthesis at the Translational Level [https://pmc.ncbi.nlm.nih.gov/articles/PMC12108789/]
- 23. Structure-Guided Design of Artificial Transcription Factor ... [https://www.sciencedirect.com/org/science/article/pii/S2161506325003250]
- 24. Genetic controllers for enhancing the evolutionary ... [https://www.nature.com/articles/s41467-025-63627-4]
- 25. Engineering a biosensor based high-throughput screening ... [https://pubmed.ncbi.nlm.nih.gov/41033575/]
- 26. Transcription Factor-Based Biosensors in High-Throughput Screening: Advances and Applications - PubMed [https://pubmed.ncbi.nlm.nih.gov/29485214/]
- 27. Utilizing a high-throughput visualization screening ... [https://www.sciencedirect.com/science/article/pii/S0956566324008121?dgcid=rss_sd_all&]
- 28. Quantitative high-throughput screening data analysis [https://pmc.ncbi.nlm.nih.gov/articles/PMC4375054/]
- 29. Engineering genetic circuits for dynamic control of central ... [https://www.sciencedirect.com/science/article/abs/pii/S109671762500093X]
- 30. Quantitative metabolomics for dynamic metabolic ... [https://www.sciencedirect.com/science/article/abs/pii/S1389172321002413]
- 31. Dynamics design of a non-natural transcription factor ... [https://www.sciencedirect.com/science/article/pii/S2405805X24000528]
- 32. Tuning the performance of a TphR-based terephthalate ... [https://www.sciencedirect.com/science/article/pii/S2214030124000191]
- 33. Engineering transcription factor-based biosensors for ... [https://microbialcellfactories.biomedcentral.com/articles/10.1186/s12934-020-01405-1]
- 34. Transcription factor-based biosensors: A molecular-guided ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8238798/]
- 35. Transcription factor-based biosensors in biotechnology [https://pmc.ncbi.nlm.nih.gov/articles/PMC4700088/]
- 36. Extending dynamic and operational range of the biosensor ... [https://www.sciencedirect.com/science/article/pii/S2405805X25000596]
- 37. A unified design allows fine-tuning of biosensor ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7593625/]
- 38. Snowprint: a predictive tool for genetic biosensor discovery [https://www.nature.com/articles/s42003-024-05849-8]
- 39. Results | HKGTC - iGEM 2025 [https://2025.igem.wiki/hkgtc/results]
- 40. Transcription Factor-Based Biosensor for Dynamic Control ... [https://www.frontiersin.org/journals/bioengineering-and-biotechnology/articles/10.3389/fbioe.2021.635265/full]
- 41. Directed Evolution of a Macrolide-Sensing Transcription ... [https://pubmed.ncbi.nlm.nih.gov/40101260/]
- 42. Directed evolution of hydrocarbon-producing enzymes [https://biotechnologyforbiofuels.biomedcentral.com/articles/10.1186/s13068-025-02689-4]
- 43. Experimental approaches to investigate biophysical ... [https://www.sciencedirect.com/science/article/pii/S1874939924000701]
- 44. Quantitative Analyses of Core Promoters Enable Precise ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4874895/]
- 45. Engineering and application of synthetic nar promoter for fine ... [https://biotechnologyforbiofuels.biomedcentral.com/articles/10.1186/s13068-018-1104-1]
- 46. Computation-guided transcription factor biosensor ... [https://www.sciencedirect.com/science/article/pii/S200103702400148X]
- 47. A hybrid approach for predicting transcription factors - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC11306938/]
- 48. Molecular docking as a tool for the discovery ... [https://www.nature.com/articles/s41598-023-40160-2]
- 49. Emerging strategies to develop novel genetically encoded ... [https://www.sciencedirect.com/science/article/abs/pii/S0167779925001593]
- 50. A domain swapping strategy to create modular ... [https://www.sciencedirect.com/science/article/abs/pii/S0734975024000399]
- 51. The Role of AI-Driven De Novo Protein Design in the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12467925/]
- 52. Computational Design of de novo Transcription Factors for ... [https://www.schrodinger.com/life-science/resources/webinar/computational-design-of-de-novo-transcription-factors-for-targeted-genetic-repression/]
- 53. Computational design of sequence-specific DNA-binding ... [https://www.nature.com/articles/s41594-025-01669-4]
- 54. Engineering chimeric signaling proteins for microbial ... [https://pubmed.ncbi.nlm.nih.gov/40903364/]
- 55. Design, modeling and in silico simulation of bacterial ... [https://www.sciencedirect.com/science/article/pii/S240584402411081X]
- 56. An Automated Cell-Free Workflow for Transcription Factor ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11494693/]
- 57. A hybrid in silico/in-cell controller that handles process ... [https://www.nature.com/articles/s41598-024-76029-1]
- 58. Development of a lysine biosensor for the dynamic regulation ... [https://microbialcellfactories.biomedcentral.com/articles/10.1186/s12934-025-02772-3]
- 59. Engineering and Application of Biosensors for Aromatic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12566513/]
- 60. Aptamer-Based Biosensors for Rapid Detection and Early ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12655300/]
- 61. Aptamer-Based Biosensors for Environmental Monitoring [https://www.frontiersin.org/journals/chemistry/articles/10.3389/fchem.2020.00434/full]
- 62. Current progress in aptamer-based sensors for the ... [https://www.sciencedirect.com/science/article/pii/S259013702400092X]
- 63. FRET Based Biosensor: Principle Applications Recent ... [https://www.mdpi.com/2075-4418/13/8/1375]
- 64. Recent Advances in Fluorescence Resonance Energy ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12025574/]
- 65. Transcription factor-based biosensors enlightened by the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4486848/]
- 66. Sensbio: an online server for biosensor design [https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-023-05201-7]
- 67. Engineering transcription factor-based biosensors for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7372789/]
- 68. Article Extended Metabolic Biosensor Design for Dynamic ... [https://www.sciencedirect.com/science/article/pii/S2589004220304922]